Fig. 4.
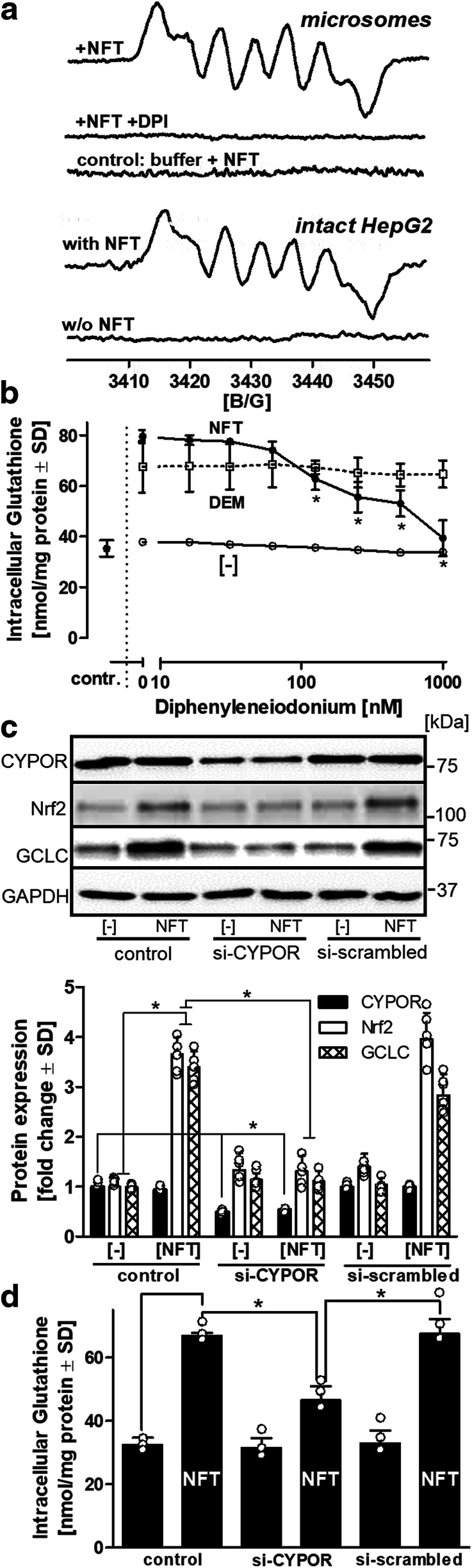
Activation of NFT. a EPR spectroscopic detection of the NFT radical anion. Microsomes (1 mg/mL protein) prepared from HepG2 in potassium phosphate buffer, pH 7.4, were supplemented with NFT (5 mM) and NADPH (10 mM) in the presence or absence of the flavoenzyme inhibitor, diphenyleneiodonium (DPI) (10 μM). Spectra were background-corrected by subtraction of the recordings obtained in absence of NFT. A total of 117 scans were recorded for microsomal preparations. Intact detached HepG2 cells (107 cells/mL) in serum-free medium were incubated in the absence or presence of NFT (5 mM). The spectrum obtained with intact HepG2 cells represents the integration of spectra from 12 scans. b HepG2 cells were exposed to DPI for 1 h, followed by the addition of NFT (40 μM) or diethyl maleate (DEM) (100 μM) for an additional 24 h. As reference, cells were exposed only to indicated concentrations of DPI. Control cells received no DPI, NFT, or DEM. Intracellular levels of glutathione were determined. c Knockdown of cytochrome P450 reductase (CYPOR). HepG2 cells were treated with siRNA (80 μL of a 10 μM stock solution per 2 × 105 cells) targeting CYPOR for 48 h. Scrambled siRNA was applied as negative control. The cells were then incubated for an additional 24 h in the presence or absence of nitrofurantoin (NFT) (40 μM). Proteins were separated by gel electrophoresis and GAPDH levels used for normalization. For each individual target, protein levels in untreated control cells were defined as unity, and alterations are expressed as fold changes compared to untreated controls. Data are means of 4 independent experiments ± SD. Individual values are indicated by dots. d In parallel, intracellular glutathione content was assessed under identical experimental conditions. Data are means of 3 independent experiments ± SD. Individual values are indicated by dots. Differences (b, d) were tested for significance by one-way ANOVA, followed by Bonferroni’s post hoc test, *p < 0.05 for (b): comparison of treatments NFT or DEM with [−] cells; (c) + (d): comparison of NFT vs. untreated control and NFT vs. siRNA knockdowns
