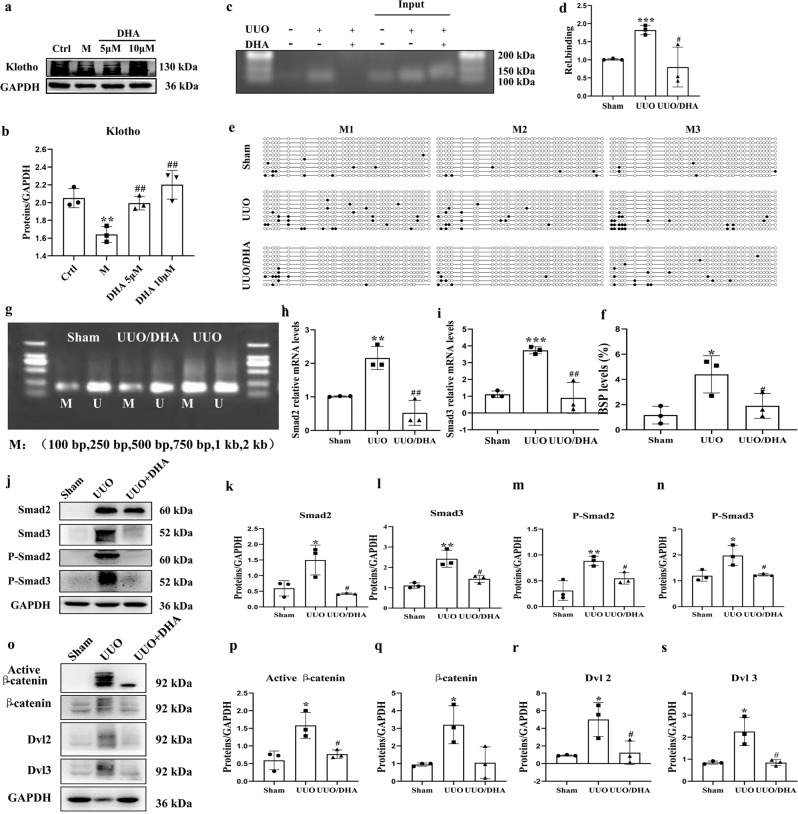
Fig. 4. DHA reverses Klotho promoter hypermethylation and inhibiting the TGF-β/Smad and Wnt/β-catenin signaling pathways in UUO mice.
a, b Expression of Klotho in primary mouse renal tubular cells (PRTCs) treated with TGF-β (10 ng/mL) in the presence or absence of DHA for 48 h as assayed by Western blotting. c, d ChIP assay. The renal tissues as indicated were immunoprecipitated with an antibody for DNMT1, and then the genomic DNA (input) and the antibody-bound DNA fragments were PCR-amplified with primers covering the DNMT1 site on the Klotho promoter. PCR products were analyzed on agarose gels and analyzed by ImageJ software. e BSP analysis of sham, UUO, and DHA-treated UUO mouse kidneys (2 weeks). Each box represents the indicated mouse kidney (M); each row of circles in the boxes represents the Klotho CpG island; each dot represents a single CpG site. Open circles indicate unmethylated (Unmethy) CpGs; filled circles indicate methylated CpGs. Each row represents a single sequenced clone (ten clones for each mouse are presented). f Quantification analysis of (e). g MSP analysis of kidney Klotho promoter methylation (Methy) from sham, UUO, and DHA-treated UUO mice (2 weeks, 3 randomly selected samples in each group). h, i Renal expression of Smad2 and Smad3 mRNA in different groups as assessed by real-time PCR. j Kidney expressions of Smad2, phosphorhospho-Smad2 Smad3, phosphorhospho-Smad3 from sham, UUO and DHA-treated UUO mice as assayed by Western blotting (three samples in each group). k–n Quantification analysis of (j). o Kidney expression of active β-catenin, β-catenin, DVL2, and DVL3 from sham, UUO and DHA-treated UUO mice as assayed by Western blot (three samples in each group). p–s Quantification analysis of (j). The results are the means ± SD of at least three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 (compared with the sham group or control group); #P < 0.05; ##P < 0.01 (compared with the UUO group or M group).