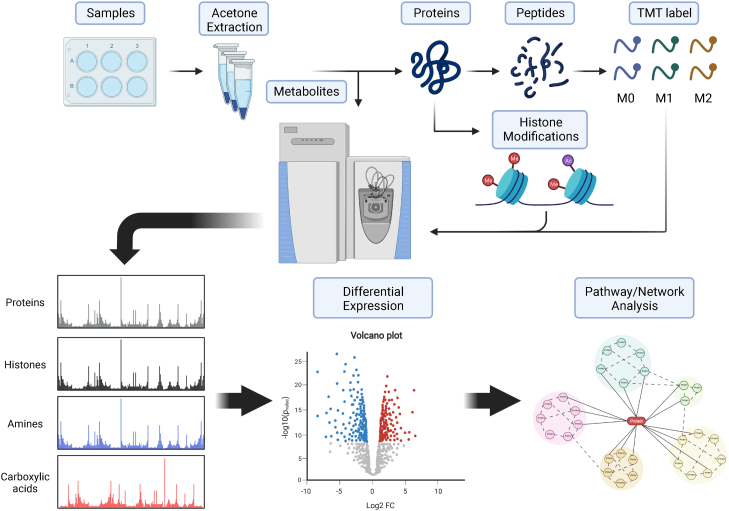
Figure 1.
Workflow for triomics for analysis of metabolites, histone modifications (epigenetics), and protein expression (phenotype). Cell lysates of M0-, M1-, and M2-macrophages (MФs) are immersed into acetone for extraction of metabolites (supernatants) and proteins (precipitates). A portion of metabolite extract is derivatized by dansylation to analyze primary amines and another portion is derivatized by O-BHA to analyze carboxylic acids by nano-LC-MS/MS. Proteins are resolved by SDS-PAGE, and gel bands containing histones (MW < 20K) are cut for in-gel digestion and histone modifications are analyzed by nano-LC-MS/MS using parallel reaction monitoring. Proteins are in-solution digested by trypsin/Lys-C. Peptides are labeled by TMT6plex. TMT6-labeled peptides are analyzed by nano-LC-MS/MS to determine protein expression changed and perturbed protein pathways. MW, molecular weight; O-BHA, O-benzylhydroxylamine; TMT, tandem mass tag.