Figure 2.
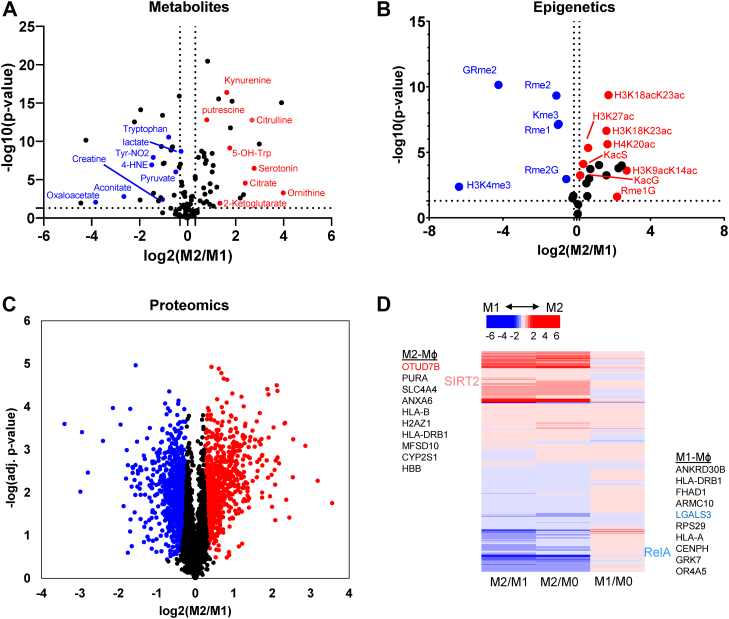
Alteration of metabolites, histone acetylation and methylation, and protein expression in M1- and M2-Mɸs.A, volcano plot of the fold change in metabolites of M1- versus M2-MФs. The x-axis is the log2 of fold changes and the y-axis is the significance of the change of each corresponding metabolite with log10 (p-value) calculated by Student's two-way t test from at least eight consecutive LC-MS/MS runs from pooled biological samples. Two separate sample preparations were prepared from three wells of 6-well plate. B, volcano plot of the fold change in histone acetylation and methylation in M1- versus M2-MФs. The plot also includes methylation of arginine, lysine amino acids or two amino acid peptides. Data were acquired from the same sample preparation as in (A). Biological and technical replicates and statistical analyses are also the same as in (A). C, volcano plot of the fold change of proteins in M1- versus M2-MФs. Protein quantification was performed with one of the two sample preparations and one additional technical replicate was applied by labeling each sample (M0-, M1-, and M2-MФs) with two TMT tags. Significance of fold changes was done with the Perseus software based on FDR set to 0.01 (1%) and s0 set to 0.32 (∼1.25× fold change cutoff). D, heat map analysis of upregulated and downregulated proteins in M2-MФ compared to M1-MФ and both relative to M0-MФ. FDR, false discovery rate; TMT, tandem mass tag.