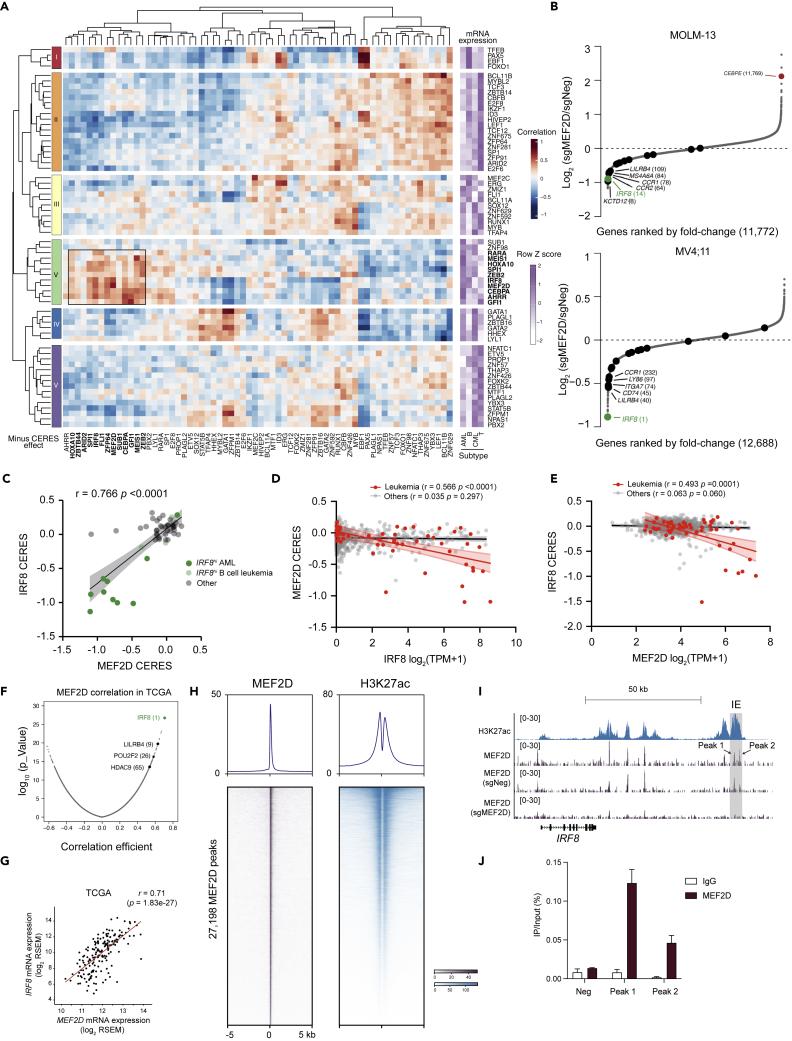
Figure 3.
MEF2D-IRF8 regulatory loop in AML
(A) Similarity matrix (left) of 66 leukemia-enriched TF dependencies in their (minus) CERES effects (row) versus mRNA expressions (column) hierarchically clustered by pair-wise Pearson correlation in 49 leukemia cell lines. Heatmap (right) implicates average expression level of TFs in different subtypes of leukemia.
(B) RNA-seq rank plot of gene expression changes in MOLM-13 (up, (Cao et al., 2021)) or MV4; 11 cells 4–5 days after transduction of two independent sgNeg or sgMEF2D. Dark dots indicate IRF8-regulated genes in Figure S3A.
(C) Scatterplot of MEF2D CERES versus IRF8 CERES in leukemia cell lines.
(D and E) Scatterplot that shows a linear correlation between IRF8’s mRNA expression and MEF2D’s CERES dependency scores (D) or MEF2D’s mRNA expression and IRF8’s CERES dependency scores (E). Each dot represents one cell line; the shaded regions indicate 95% confidence interval for the linear regression model.
(F) Pearson correlation analysis of MEF2D mRNA expression with other genes in 173 AML patients. Data retrieved from TCGA dataset.
(G) Scatterplot of MEF2D and IRF8 expression in 173 AML patients.
(H) Meta-profile (top) and density plot (bottom) of MEF2D CUT&RUN and H3K27ac ChIP-seq peaks in MOLM-13 cells. Peaks are ranked by MEF2D CUT&RUN tag counts.
(I) Gene tracks of H3K27ac and MEF2D enrichment at IRF8 locus in MOLM-13 cells. Shadowed in gray is the AML-specific IRF8 enhancer (IE). (J) CUT&RUN-qPCR against MEF2D on IRF8 locus (n = 3).