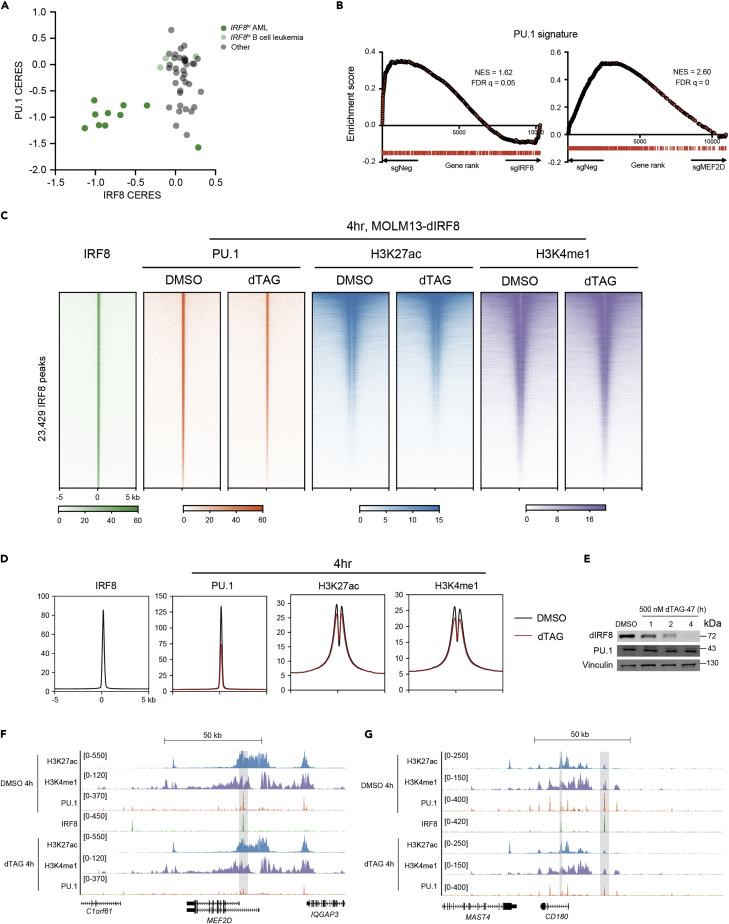
Figure 4.
IRF8 supports PU.1’s chromatin occupancy and transcriptional output
(A) Scatterplot of IRF8 CERES versus PU.1 CERES in leukemia cell lines.
(B) GSEA analysis of IRF8-KO (left) and MEF2D-KO (right) RNA-seq data in MOLM-13 cells. PU.1 signature scatterplot of MEF2D CERES versus IRF8 CERES in leukemia cell lines, where the PU.1 signature is defined as the top 500 downregulated genes on PU.1 depletion. Normalized enrichment score (NES) and false discovery rate (FDR) q value are shown.
(C and D) Density plot (C) and meta-profiles (D) of PU.1, H3K27ac and H3K4me1 ChIP-seq signals at IRF8 binding sites in MOLM13-dIRF8 cells (Cao et al., 2021) on 4 h treatment of DMSO or dTAG that rapidly eliminates IRF8 protein. Peaks are ranked by IRF8 ChIP-seq tag counts (n = 3).
(E) Immunoblotting of IRF8, PU.1 and GAPDH in whole-cell lysates of MOLM13-dIRF8 cells treated with 500nM dTAG-47 for 4h.
(F and G) Gene tracks of ChIP-seq signal from Figures 4C and 4D at MEF2D (E) or CD180 (F) locus.