Figure 5.
MEF2D and IRF8 are upregulated in AML carrying KMT2A-r through enhancer reactivation
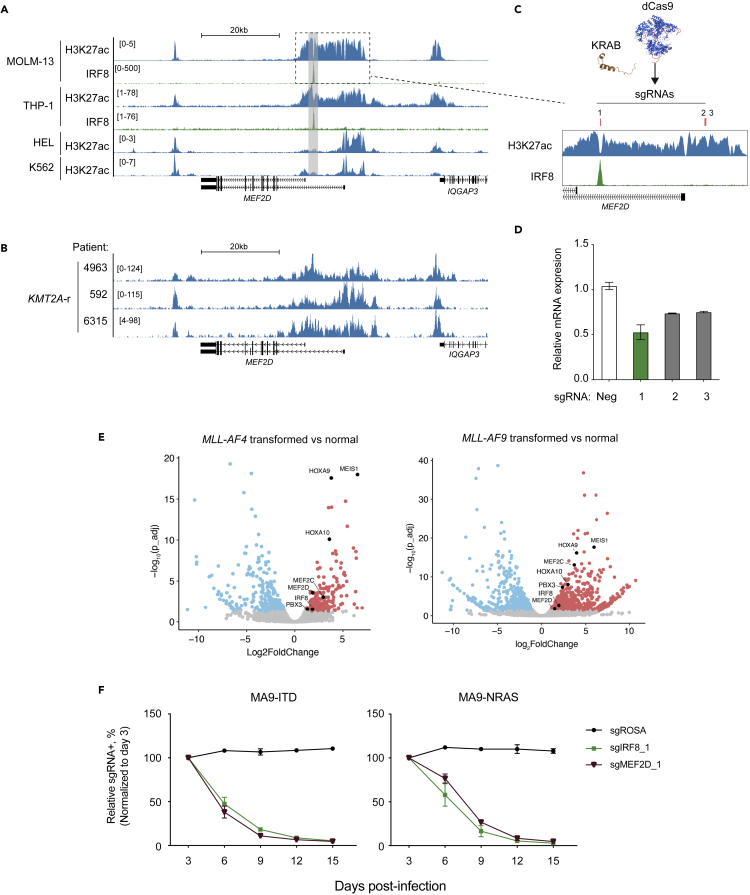
(A) Gene tracks of H3K27ac and IRF8 ChIP signals in MOLM-13, THP-1, HEL or K562 cells.
(B) Gene tracks of H3K27ac CUT&RUN signals in primary AML cells, with the key oncogenic mutations labeled on the left.
(C) Schematic of CRISPRi (KRAB-dCas9)-mediated epigenomic silencing at MEF2D locus. Red bars indicate sgRNA positions.
(D) RT-qPCR analysis of mRNA expression of MEF2D in MOLM-13 cells stably expressing dCas9-KRAB and transduced with indicated sgRNAs in Figure 5B harvested on 4 days post-infection. Relative mRNA levels were normalized to GAPDH levels. Plotted are the mean ± SEM (n = 3 for negative controls, n = 2 for MEF2D sgRNAs).
(E) Volcano plots of RNA-seq data in MA4- (left) or MA9- (right) transformed human CD34+ HSPCs versus normal HSPCs isolated from BM, where KMT2A translocation was induced via CRISPR-Cas9 (Secker et al., 2020).
(F) Competition-based proliferation assays in MA9-FLT3ITD (left) or MA9-NRASG12D (right) cells, which were generated by retroviral transduction of KMT2A-AF9 followed by FLT3 FLT3ITD and NRASG12D into human CD34+ HSPC cells, respectively (Wei et al., 2008; Wunderlich et al., 2013) (n = 3, mean ± SEM).