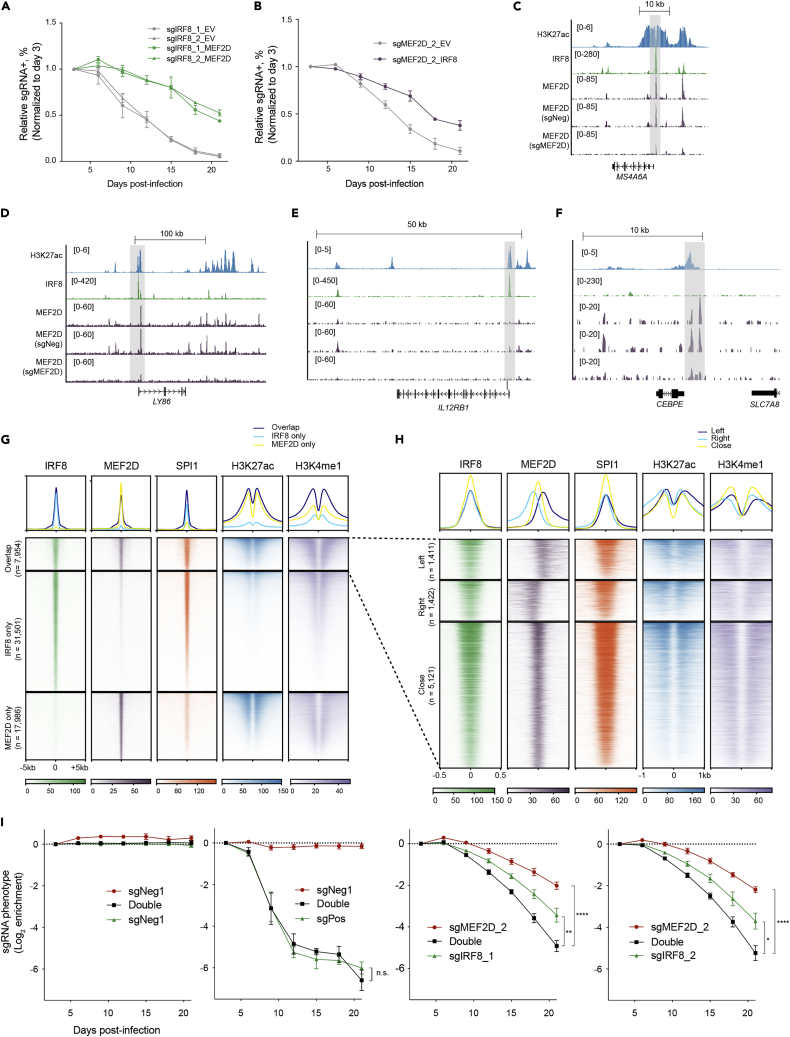
Figure 6.
Divergent function of MEF2D- and IRF8- regulated programs in supporting AML
(A) Competition-based proliferation assays in THP-1 cells transduced with EV or MEF2D cDNA (n = 3–4, mean ± SEM).
(B) Competition-based proliferation assays in THP-1 cells transduced with EV or IRF8 cDNA (n = 3–4, mean ± SEM).
(C–F) Exemplary tracks of genes where MEF2D and IRF8 exact overlap (C), bind in proximity with each other (D), or only IRF8 (E) or MEF2D (F) are enriched.
(G) Meta-profile (top) and density plot (bottom) of IRF8, MEF2D, SPI1, H3K27ac and H3K4me1 enrichment at IRF8-MEF2D co-bound, IRF8-only and MEF2D-regions in MOLM-13 cells. Peaks are ranked by IRF8 or MEF2D tag counts.
(H) Meta-profile (top) and density plot (bottom) depicting detailed classification of IRF8-MEF2D co-bound regions: close, IRF8 and MEF2D peaks have at least 1bp overlap; left, IRF8 binds upstream and MEF2D downstream of the chromosome; right, IRF8 binds downstream and MEF2D upstream of the chromosome. IRF8, MEF2D, SPI1, H3K27ac and H3K4me1 enrichment were plotted. Peaks are ranked by IRF8 tag counts and centered by IRF8 peaks.
(I) Competition-based proliferation assays in THP-1 cells simultaneously transduced with two indicated sgRNAs linked with GFP or mCherry, which produced a mixed population of uninfected, single, and double infected cells. Relative fluorescent (GFP+/cherry+, GFP+/cherry-, GFP-/cherry+) proportion of a single or combinatorial sgRNA was normalized with uninfected cells as an internal control, and the normalized ratios were further compared with that on day 3 to calculate the enrichment of each population (y-axis). sgPos + sgNeg group showed the genome editing was not compromised when two sgRNAs were applied, and no additive/synergistic effect was seen in this combination. (n = 3 for infection with positive and negative controls, n = 5–8 for sgIRF8+ sgMEF2D, mean ± SEM. n.s, not significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.001, two-tailed Welch’s unpaired t-test).