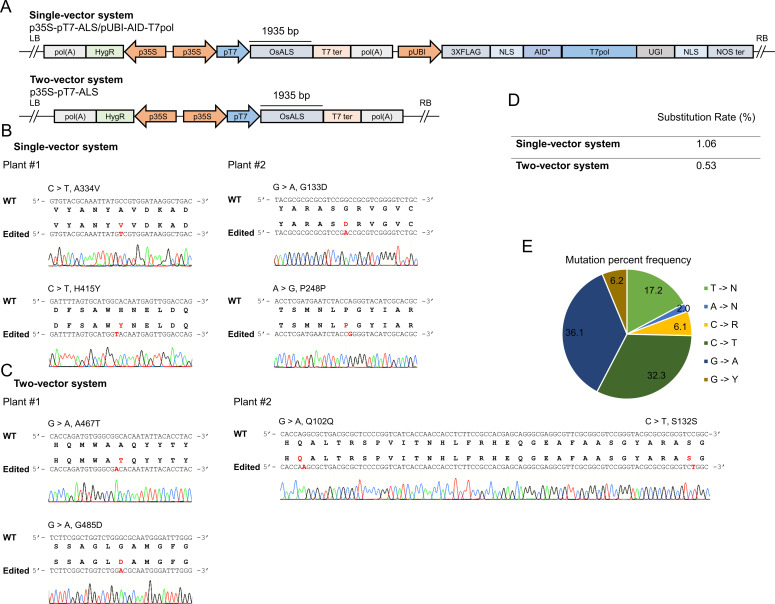
Figure 2. Targeted base editing in stable rice lines.
(A) Design of the plasmid used for Agrobacterium-mediated transformation of callus. In single-vector editing, the target and editor were cloned in the same vector and transformed into rice. In the two-vector system, OsALS was inserted into the vector under the control of the T7 promoter and co-transformed with pEditor. (B) Sanger sequencing analysis of regenerated rice shoots transformed with the single-vector or two-vector system. (C) 10 reads were analyzed per plant. Most seedlings showed C > T and G > A substitutions. (D) Amplicon deep sequencing analysis of acetolactate synthase (ALS) targeted with the single-vector or two-vector system. p35S:ALS without the base editor was used as a mock control. The amplicons were sequenced using PacBio technology. The substitution rate was calculated after subtracting the mock-treated sample reads. (E) Mutation percent frequency represents the distribution of base edits of all types found in the ALS samples with p35S-AID-T7 RNAP. The data are the means of the results for the single- and two-vector systems.
Source data are available for this figure.