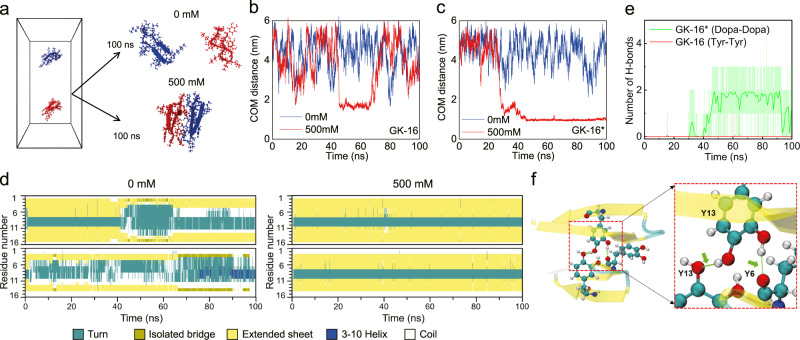
Fig. 3. MD simulations of the pair interactions of GK-16 and GK-16* peptides.
a Simulation models; left: simulation box with initially separated peptides; right: the final snapshots of GK-16* under 0 mM or 500 mM ionic strength at 100 ns. Water molecules and ions are not shown for clarity. The representative temporal evolutions of COM distances between the pairs of peptides with 0 mM or 500 mM ionic strength, for GK-16 (b) and GK-16* (c). d Residue maps of the secondary structures as a function of time for GK-16* under 0 mM and 500 mM ionic strength in simulations. e The temporal evolutions of the number of H-bonds formed between Dopa residues in GK-16* and Tyr residues in GK-16 (transparent line) and the corresponding average over 1 ns (solid line) during the interaction process. f Representative configurations of the associated GK-16*; the right panel shows a magnified view of the bidentate H-bonds, green arrows indicating the H-bonds. Atoms in Dopa residues are colored as follows: hydrogen, white; oxygen, red; carbon, cyan; nitrogen, blue.