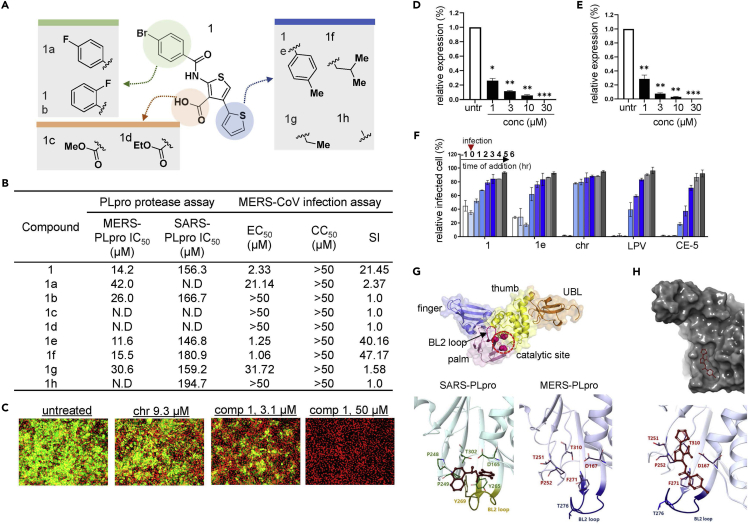
Figure 3.
Thiophene and its analogs prevent both MERS-CoV and SARS-CoV proliferation
(A) Structural presentation of thiophene (1) and its analogs.
(B) Summary of the potency of each analog. The potency of each thiophene analog is presented as the 50% inhibitory concentration (IC50) value measured using the fluorescence-based MERS-PLpro and SARS-PLpro protease assay. The 50% effective concentration (EC50), 50% cytotoxicity concentration (CC50), and selective index (SI; EC50 divided by CC50) were determined by the immunofluorescence-based infection assay using MERS-CoV-infected Vero cells.
(C) Representative images from the MERS-CoV infection assay. Vero cells were infected with MERS-CoV followed by treatment with compound 1 or the control compound chloroquine (chr) for two days at concentrations ranging from 0.0977 to 50 μM, and images were taken on day 2. Green signals indicate cells infected with MERS-CoV and red signals indicate live Vero cells.
(D and E) RNA quantification of the viral upE gene in MERS-CoV-infected cells after treatment with compound 1 (D) or analog 1a (E). Each data point represents the mean of quadruple assays with ±SEM. ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005 compared to untreated control by Student’s t test. untr: untreated control.
(F) Time-of-addition experiment using compound 1. The effect of compound 1 on the early or late stage of infection was determined by adding the compound prior to infection (−1 h), at the time of infection (0), and post-infection (1-6 h) and by subsequently quantifying the upE gene in virus-infected Vero cells. Each data point represents the mean of duplicate assays with ±SD. LPV: lopinavir; CE-5: chloropyridine ester.
(G) Structure of PLpro. PLpro is mainly composed of finger, thumb, palm, and ubiquitin-like (UBL) domains. The BL2 loop and catalytic site important for molecular interaction are included in the palm domain. The BL2 loop and important residues for SARS-PLpro (left) and MERS-PLpro (right) are compared. Crystal structure of SARS-PLpro with its inhibitor (PDB: 3E9S) is adapted from reference (Ratia et al., 2008).
(H) Image of molecular docking of compound 1 into MERS-PLpro (PDB: 4RF1). Docking was performed using the XP Glide docking tool of the Schrodinger platform (v. 2019-2).