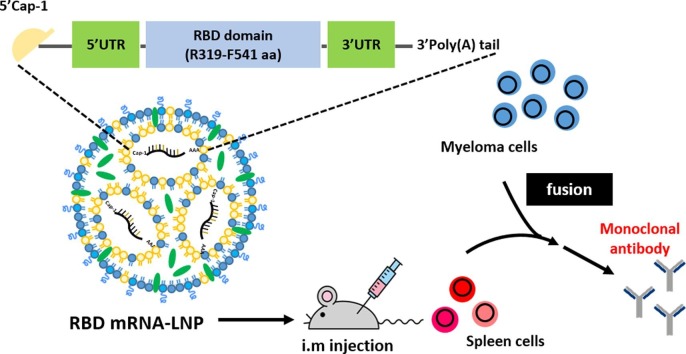
Graphical abstract
Keywords: SARS-CoV-2, COVID-19, Lipid nanoparticle (LNP), Modified mRNA, Monoclonal antibodies
Abstract
Throughout the COVID-19 pandemic, many prophylactic and therapeutic drugs have been evaluated and introduced. Among these treatments, monoclonal antibodies (mAbs) that bind to and neutralize SARS-CoV-2 virus have been applied as complementary and alternative treatments to vaccines. Although different methodologies have been utilized to produce mAbs, traditional hybridoma fusion technology is still commonly used for this purpose due to its unmatched performance record. In this study, we coupled the hybridoma fusion strategy with mRNA-lipid nanoparticle (LNP) immunization. This time-saving approach can circumvent biological and technical hurdles, such as difficult-to-express membrane proteins, antigen instability, and the lack of posttranslational modifications on recombinant antigens. We used mRNA-LNP immunization and hybridoma fusion technology to generate mAbs against the receptor binding domain (RBD) of SARS-CoV-2 spike (S) protein. Compared with traditional protein-based immunization approaches, inoculation of mice with RBD mRNA-LNP induced higher titers of serum antibodies and markedly increased serum neutralizing activity. The mAbs we obtained can bind to SARS-CoV-2 RBDs from several variants. Notably, RBD-mAb-3 displayed particularly high binding affinities and neutralizing potencies against both Alpha and Delta variants. In addition to introducing specific mAbs against SARS-CoV-2, our data generally demonstrate that mRNA-LNP immunization may be useful to quickly generate highly functional mAbs against emerging infectious diseases.
1. Introduction
As the world continues to grapple with the coronavirus COVID-19 pandemic (Zhu et al., 2019, Sun et al., 2020), monoclonal antibodies (mAbs) have become increasingly used in applications such as basic research, diagnosis, therapeutics for SARS-CoV-2 infection (Hwang et al., 2022, Chapman et al., 2021). Fortunately, the hybridoma technology developed by Georges Kohler and Cesar Milstein in 1975 has made it possible to obtain large numbers of mAbs for these purposes (Köhler and Milstein, 1975). Despite its widespread application, hybridoma technology still suffers from some limitations. For example, the conventional methodology used for hybridoma generation involves multiple injections of a protein antigen with or without adjuvant (Chiarella and Fazio, 2008). It is sometimes a major challenge to prepare high-quality protein antigen for immunization, yet this step is necessary to generate high-precision mAbs that are able to recognize the native viral antigen (Takeda et al., 2015). Purification of protein antigens is also a time consuming and labor-intensive process, as optimized protocols must be created for each target (Holzlöhner and Hanack, 2017). Moreover, it is technically difficult to express or purify certain antigens, such as transmembrane, glycosylated, toxic, or unstable proteins (Parray et al., 2020, Zuo et al., 2005). Once the antigen is produced, the purified recombinant proteins are often mixed into adjuvant formulations that might alter the native protein conformations and lead to unexpected immune responses and undesired consequences in the immunized animals (Parray et al., 2020). In addition, it is well known that maintenance of structural integrity of immunogens is a critical concern for the induction of functional mAbs (Liu et al., 2016). For all of these reasons, the use of protein antigens is often inadequate, and an alternative approach of expressing intact immunogens in vivo may be better suited for inducing mAbs with desired biological activities.
In recent years, the ability to produce mRNA in a cell-free environment by in vitro transcription (IVT) has provided a realistic opportunity to generate any protein of interest in cells or animals (Maruggi et al., 2019). However, the use of mRNA to induce protein production in animals is complicated by its susceptibility to degradation by nucleases, inherent instability, stimulation of excessive inflammatory responses and inefficient in vivo delivery (Hou et al., 2021, Wang et al., 2021). During the last two decades, intensive research has been carried out to develop effective strategies for stabilization and delivery mRNA (Jackson et al., 2020, Pardi et al., 2018). As a result of these efforts, co-formulation into lipid nanoparticles (LNPs) has become one of the most successful and commonly used methods for mRNA delivery in vivo (Ickenstein and Garidel, 2019). Because of its reliability, mRNA-LNP is being extensively utilized in a broad range of new and potential treatments, such as regenerative medicines, vaccines, immunotherapies, and gene editing applications (Baptista et al., 2021).
In this study, we designed and developed an LNP-encapsulated nucleoside-modified SARS-CoV-2 RBD mRNA (amino acids [aa] 319–541) with IgG kappa chain leader sequence for protein secretion (called RBD mRNA-LNP). The expression levels and secretory efficiencies of RBD mRNA-LNP were initially evaluated in HEK293T cells by Western blotting and ELISA. We then immunized mice with RBD mRNA-LNP over a period of 7 weeks and found that the animals had higher serum antibody titers than mice vaccinated with RBD protein in adjuvant over a period of 10 weeks. After using the mice for hybridoma production, we identified six different IgG mAbs, demonstrating the applicability of this novel immunization approach for mAb generation. Each of the six mAbs was evaluated in terms of its target binding affinity and neutralization potential using ELISA and pseudovirus neutralization assays. The mAbs were all found to exhibit specific binding to SARS-CoV-2 RBD protein and at least some of the tested variants. Among the six mAbs, RBD-mAb-3 showed remarkably potent neutralization against both Alpha and Delta variants (Alpha, IC50 = 10.99 ± 1.45 ng/ml and Delta, IC50 = 11.12 ± 1.09 ng/ml). Binding of RBD-mAb-3 to S protein RBD was inhibited by mutations at sites Y453A and Q474A. Together, these data suggest that mRNA-LNP immunization has the potential to overcome or bypass many current challenges in mAbs development, allowing fast turnaround times that can benefit efforts to monitor or treat emerging infectious diseases.
2. Results
2.1. Design and characterization of the RBD mRNA-LNP immunogen for SARS-CoV-2
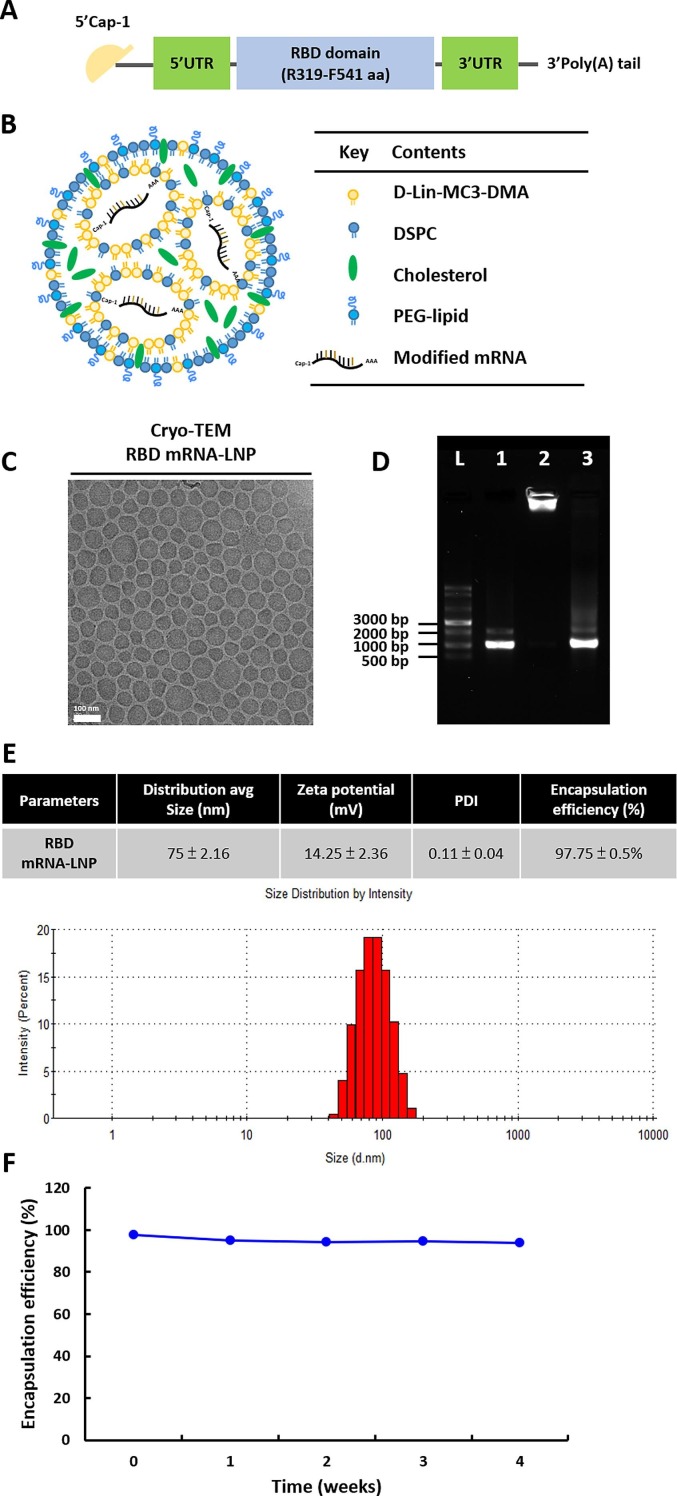
Lipid nanoparticles (LNPs) are one of the most commonly used tools for in vivo delivery of mRNA (Ickenstein and Garidel, 2019). To develop an mRNA-LNP to deliver the RBD of SARS-CoV-2 as an immunogen, the RBD of SARS-CoV-2 (amino acids [aa] 319–541) was produced by IVT from a template based on the published sequence of the virus (Fig. 1 A). This synthetic mRNA harbored an optimal Cap-1 structure and a signal peptide sequence from human IgG kappa chain (IgGκ). N1-methyl-pseudouridine (m1Ψ) was substituted for uridine throughout the mRNA sequence to increase mRNA stability and translation (Parr et al., 2020). The LNP was composed of four types of lipids, including D-Lin-MC3-DMA, DSPC, cholesterol and PEG-lipid at molar ratios of 50:10:38.5:1.5 (Fig. 1B). After production, the RBD mRNA-LNP samples were observed by cryo-electron microscopy (Fig. 1C), which revealed the formulation was comprised of spherical and homogenously distributed LNPs without aggregation. As shown in Fig. 1D, agarose gel retardation was performed in presence and absence of Triton X-100. mRNA encapsulated in the LNP particles (lane 2) showed no free mRNA band, suggesting that the encapsulation of mRNA into the LNPs was complete. Following the addition of Triton X-100 to disrupt the lipid membrane (lane 3), the mRNA was released from the LNPs but did not exhibit any degradation. Since the mRNA released from the disrupted LNP was at the same molecular weight as the naked mRNA, we concluded that the integrity of mRNA was not altered by encapsulation. The particle size distribution of RBD mRNA-LNP ranged from about 70 to 90 nm, and PDI was less than 0.15, indicating a mono-dispersed size distribution (Fig. 1E). Of note, the positive surface charge of LNPs may allow for their interaction with negatively charged plasma membrane to facilitate cellular uptake. In our formulation, the particle size distribution and surface charge were suitable for cellular uptake by endocytosis pathways. To further explore the stability of mRNA-LNP, the encapsulation efficiency (EE%) was measured after storage at 4 °C for up to 4 weeks (Fig. 1F). Remarkably, the EE% was not changed over the duration of the experiment. These results suggested the RBD mRNA-LNP is stable when stored at 4 °C over a period of 4 weeks post-formulation.
Fig. 1.
Design and physicochemical characterization of the modified RBD mRNA-LNP used for immunization. (A) Schematic illustration of SARS-CoV-2 RBD mRNA; comprises a 5′ cap, 5′ UTR, signal peptide, antigen (RBD of the spike protein), 3′ UTR, and 3′ poly(A) tail. (B) Schematic representation of the LNP-encapsulated RBD mRNA. (C) Cryo-TEM imaging was performed on RBD mRNA-loaded LNPs; the LNPs consisted of D-Lin-MC3-DMA:DSPC:Cholesterol:DMG-PEG 2000 at 50:10:38.5:1.5 molar ratios. Scale bar, 100 nm. (D) Agarose gel electrophoresis of RBD mRNA-LNP was performed with and without 1% Triton X-100 to assess mRNA integrity. Naked mRNA was used as a control. L = single-stranded RNA ladder, 1 = RBD mRNA alone, 2 = RBD mRNA-LNP (in 1X TE), 3 = disrupted RBD mRNA-LNP (in 1% Trition X-100). (E) Average size, zeta potential and PDI distribution were measured with dynamic light scattering (DLS); Encapsulation efficiency (EE%) was determined with a Ribogreen assay kit.
2.2. In vitro characterization of RBD mRNA or RBD mRNA-LNP expression
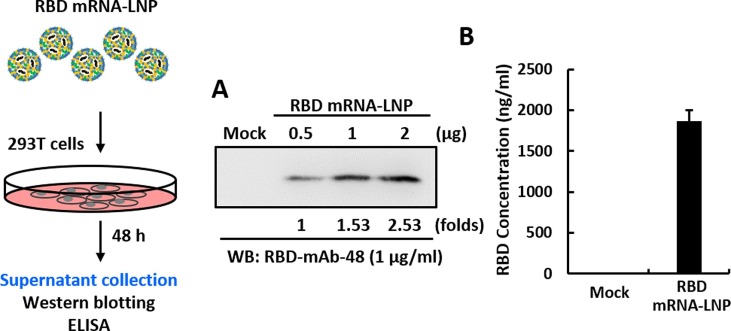
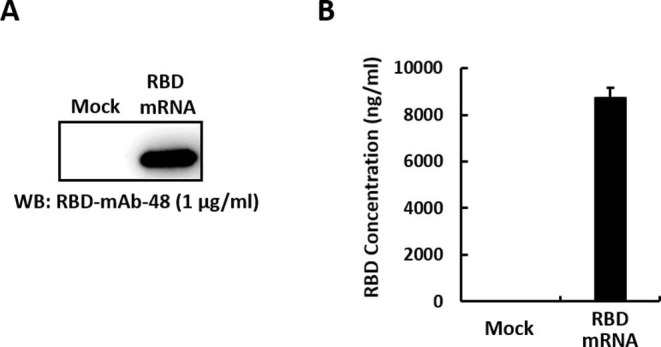
To assess cell entry, cargo escape and translation of the transported mRNA molecules, RBD protein expression was measured after transfecting HEK293T cells with RBD mRNA-LNP. Two days after transfection, RBD protein secreted into the medium was analyzed by Western blotting (Fig. 2 A) and ELISA (Fig. 2B). Negative controls included media from non-transfected cells. The results demonstrated that the RBD could be efficiently produced and secreted by HEK293T cells into the culture supernatant. Quantitative densitometric analysis of the Western blot showed that mRNA-LNP transfection induces RBD protein expression in a dose-dependent manner. In addition, we evaluated the translation efficiency of RBD mRNA after lipofectamine-mediated transfection into cell cultures (Supplementary Fig. 1).
Fig. 2.
In vitro characterization of immunogen expressed from RBD mRNA-LNP. Schematic of the RBD mRNA-LNP transduction protocol for HEK293T cells. (A, B) RBD protein expression level was measured in HEK293T cells transfected with RBD mRNA-LNP by Western blotting (A) and ELISA (B), using an anti-RBD antibody. Mock transfected HEK293T cells served as a negative control.
2.3. Immunization with RBD mRNA-LNP elicits a robust antigen-specific immune response in mice
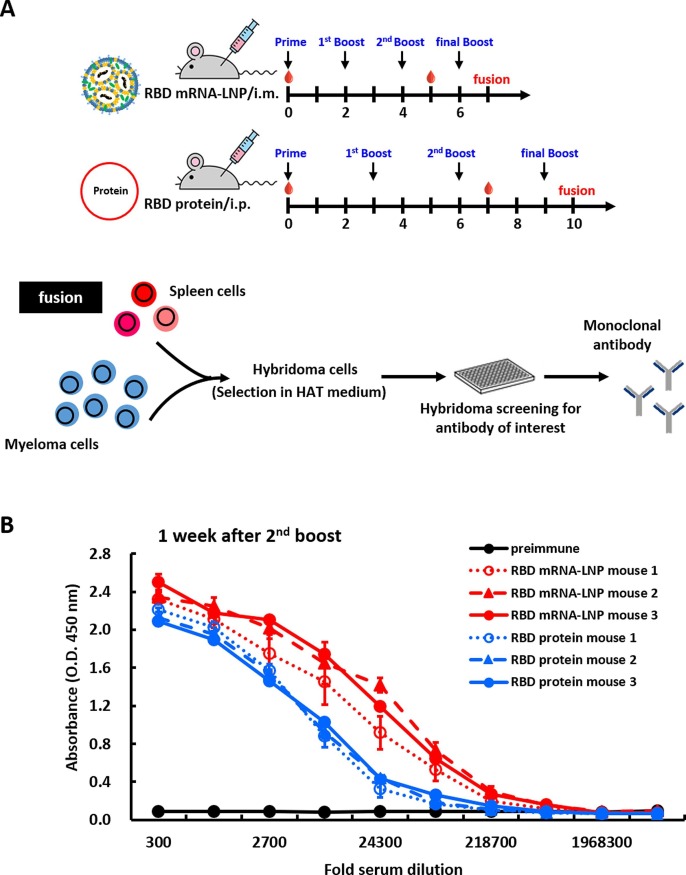
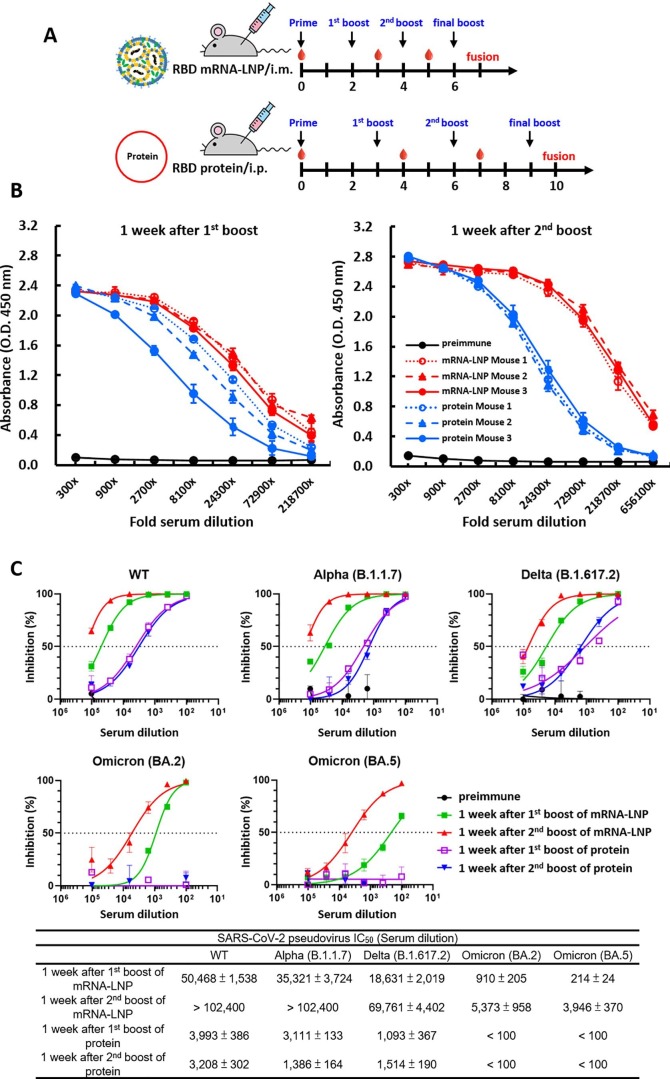
Next, we wanted to compare the strengths and durations of antibody responses between mice immunized with RBD mRNA-LNP and those subjected to a traditional protein-based approach. BALB/c mice were immunized via intramuscular injection with RBD mRNA-LNP at 2-week intervals or received intraperitoneal injection with recombinant RBD protein in CFA-IFA adjuvant at 3-week intervals. A schematic representation of the immunization schedule is shown in Fig. 3 A. Pre-immune and immune serum were respectively collected before the priming injection (week 0) and one week after the second booster. Subsequently, the RBD-binding antibody titers of the serum samples were evaluated by ELISA. Sera from mice immunized with the RBD mRNA-LNP exhibited significantly higher titers against RBD protein, as compared to the mice immunized with the protein and adjuvant (Fig. 3B). Next, we confirmed the functionality of our approach by showing that antibody titers in mRNA-LNP-immunized mice were indeed higher than those of protein-immunized mice over time. Mouse immunizations were conducted as shown in Supplementary Fig. 3A, and ELISA was used to examine the binding ability of serum samples collected at one week after first or second booster with recombinant RBD protein (Supplementary Fig. 3B). The results showed that no matter whether sera were obtained at early or late time points, the mice immunized with the RBD mRNA-LNP showed higher antibody titers against RBD protein, as compared to mice immunized with protein and adjuvant. We also compared the neutralizing abilities of these immunized mouse sera against WT and mutant SARS-CoV2 variants as shown in Supplementary Fig. 3C. Either one week after the first or second booster, mice immunized with RBD mRNA-LNP exhibited higher neutralizing activities toward different SARS-CoV-2 pseudoviruses, including WT, Alpha, Delta and Omicron (BA.2 and BA.5) variants, as compared to the mice immunized with protein and adjuvant. These results suggest that two doses of RBD mRNA-LNP immunization induced a strong immune response in mice against the wild-type SARS-CoV-2. Furthermore, the second booster dose was more immunogenic in mice, suggesting that this regimen is highly suitable for production of mAbs against SARS-CoV-2 and its variants (Kumari et al., 2022).
Fig. 3.
Immunization with RBD mRNA-LNP elicits a robust antigen-specific immune response in mice. (A) Schematic of immunization regimens in mice. Female BALB/c mice (n = 3 per group) were immunized by three i.m. injections of RBD mRNA-LNP at 2-week intervals or by three i.p. injections of recombinant RBD protein at 3-week intervals. Red droplets indicate blood draws. After the final booster, the spleens were harvested and fused with NS1 cells to generate hybridomas, which were screened for RBD-mAbs. (B) ELISA was used to determine the titers of preimmune and hyperimmune sera against RBD mRNA-LNP or RBD protein after three immunizations. Mouse sera were collected before immunization and after the third immunization and incubated with RBD protein-coated plates. The absorbance values were read at O.D. 450 nm.
2.4. Generation and characterization of monoclonal antibodies
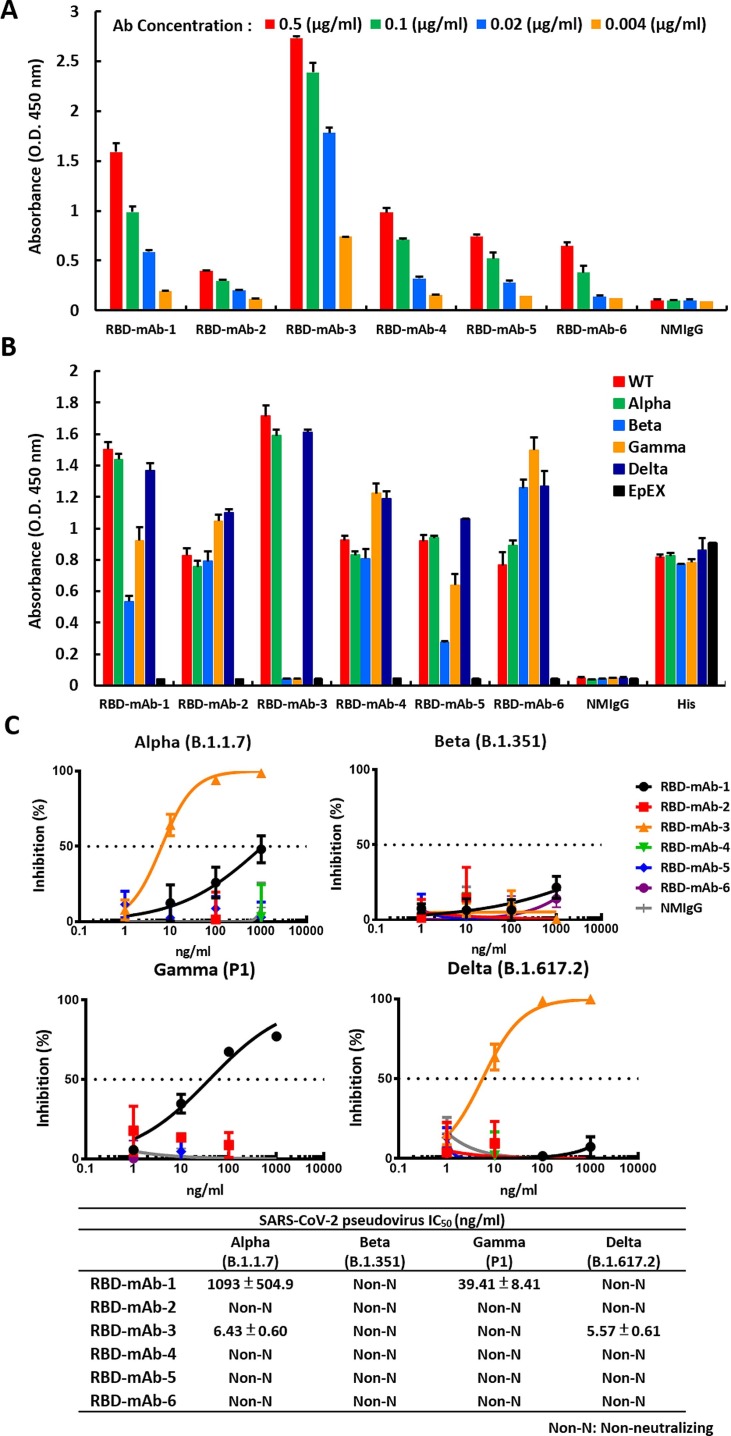
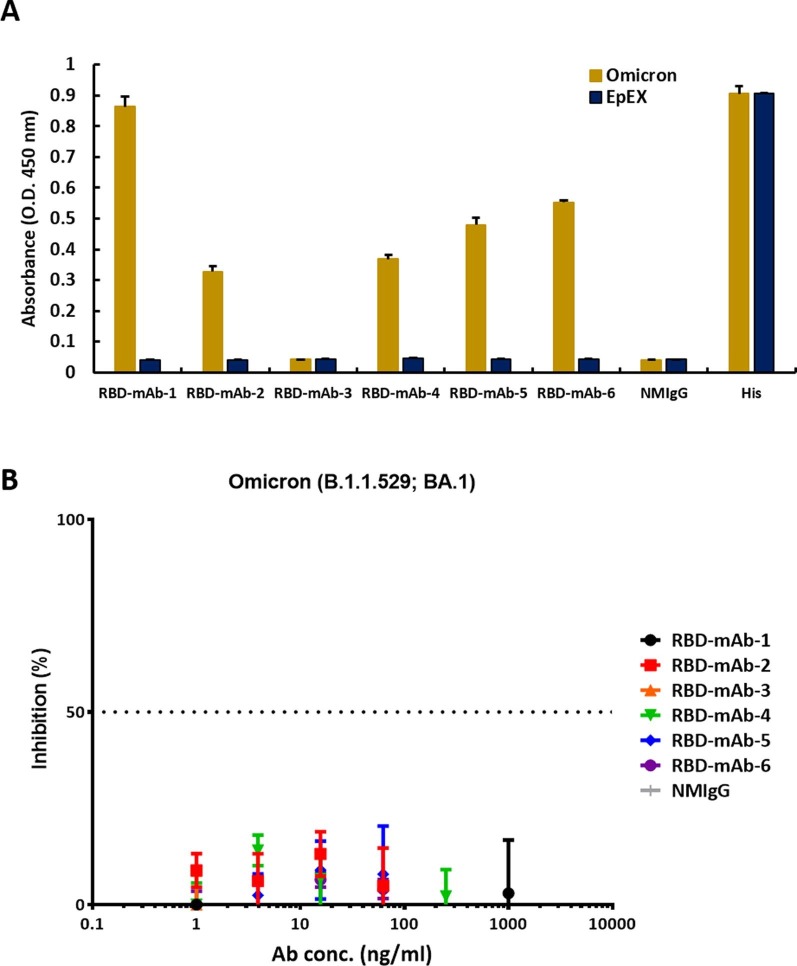
RBD mRNA-LNP is highly immunogenic in mice and is suitable for subsequent production of mAbs against RBD. As such, the RBD mRNA-LNP-immunized mice were used for cell fusion. One week after the final injection, splenocytes were harvested from the mice and fused with NS1/1-Ab4-1 (NS-1) mouse myeloma cells. The resulting fused hybrid cells were cultured and selected in HAT tissue culture media. Supernatants of hybridoma clones were screened by ELISA for binding to recombinant RBD protein. As shown in Fig. 4 A, six hybridoma culture supernatants containing anti-RBD IgG1 (κ light chain) showed specific dose-dependent binding to RBD protein. Nevertheless, like many other RNA viruses, SARS-CoV-2 has a high mutation rate, and some mutations may cause new variants to be more transmissible and virulent than the original SARS-CoV-2 wild-type strain, reported in late 2019 (Harvey et al., 2021). Over the course of the pandemic, five SARS-CoV-2 lineages have been designated by the World Health Organization as variants of concern (VOCs), including Alpha (B.1.1.7), Beta (B.1.351), Gamma (P1) and Delta (B.1.617.2) and the recent Omicron variant (B.1.1.529). Therefore, we also tested the binding capabilities of our hybridoma culture supernatants to RBDs with mutations corresponding to those found in the five VOCs (Fig. 4B and Supplementary Fig. 2A). RBD-mAb-1 and RBD-mAb-5 recognized wild-type RBD and RBD variants with mostly similar binding activities, but showed relatively weaker binding to the Beta RBD variant. Notably, RBD-mAb-1 showed efficient binding to the Omicron RBD variant, while RBD-mAb-5 had much weaker binding activity for the Omicron RBD variant. RBD-mAb-2, RBD-mAb-4 and RBD-mAb-6 bound well to wild-type RBD and most RBD variants, but their binding to Omicron RBD was relatively weak. RBD-mAb-3 bound strongly to wild-type RBD, Alpha RBD, and Delta RBD, but had no detectable binding to the Beta, Gamma and Omicron variants. In addition to evaluating the binding of hybridoma supernatants to recombinant proteins, we examined the neutralizing potencies of our six hybridoma culture supernatants toward pseudoviruses of the five SARS-CoV-2 VOCs. Consistent with the ELISA data showing that RBD-mAb-3 bound to Alpha and Delta RBD variants, the pseudovirus neutralization assays revealed that RBD-mAb-3 exhibited high neutralizing abilities for Alpha and Delta variants, with IC50 values of less than 6.43 ng/ml (Fig. 4C). While this selected hybridoma clone was effective against the virulent Delta strain, it did not effectively neutralize the Omicron pseudovirus (Supplementary Fig. 2B). We have also evaluated he Regeneron monoclonal antibodies REGN10933 (Casirivamab) and REGN10987 (Imdevimab), the Eli Lilly cocktail [consisting of LY-CoV555 (Bamlanivimab) and LY-CoV016 (Etesevimab)], as well as RBD-mAb-1 and RBD-mAb-3 for the ability to neutralize SARS-CoV-2 variants of concern (Table 1 ) (Hwang et al., 2022, Du et al., 2022, Lusvarghi et al., 2022). REGN10933 (Casirivamab) potently neutralized Alpha and Delta, but had no detectable activity against Beta, Gamma and Omicron. REGN10987 (Imdevimab) potently neutralized earlier viruses but lacked activity against Omicron virus. LY-CoV555 (Bamlanivimab) neutralized Alpha virus, but was inactive against other variants. LY-CoV016 (Etesevimab) potently neutralized Alpha and Delta viruses but lacked activity against Beta, Gamma and Omicron viruses. Moreover, our newly discovered mAb RBD-mAb-1 had weak activity against Alpha and Gamma viruses but was inactive against the Beta, Delta and Omicron viruses. RBD-mAb-3 exhibited strong neutralizing activity against the Alpha and Delta viruses. Based on these findings, the Omicron variant with more than 30 mutations in its spike protein is likely to escape neutralization by most therapeutic antibodies.
Fig. 4.
Generation and characterization of mAbs from hybridomas. (A) Hybridomas were generated, and serial dilutions of the cell culture supernatants were screened for RBD WT-His binding ability by ELISA. (B) The binding affinities of hybridoma culture supernatants were also assessed for RBDs of different SARS-CoV-2 variants by ELISA. EpEX protein served as a negative control. (C) Neutralization assays were performed with hybridoma culture supernatants targeting four SARS-CoV-2 VOC pseudoviruses. Assays were performed in triplicate; each point represents the mean ± SEM. IC50 values were calculated with GraphPad Prism software.
Table 1.
Neutralization of SARS-CoV-2 variants by therapeutic mAbs.
| Antibody name | Alpha (B.1.1.7) | Beta (B.1.351) | Gamma (P.1) | Delta (B.1.617.2) | Omicron (B.1.1.529) |
|---|---|---|---|---|---|
| REGN10933 (Casirivimab) | +++ | – | – | +++ | – |
| REGN10987 (Imdevimab) | +++ | +++ | +++ | +++ | – |
| LYCoV-555 (Bamlanivimab) | +++ | – | – | – | – |
| LYCoV-016 (Etesevimab) | + | – | – | +++ | – |
| RBDmAb-1 | + | – | ++ | – | – |
| RBDmAb-3 | +++ | – | – | +++ | – |
The neutralization abilities of therapeutic mAbs against SARS-CoV-2 variants, including B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), B.1.617.2 (Delta) and B.1.1.529 (Omicron). Symbols indicate the range of half maximal inhibitory concentration (IC50) values toward authentic or pseudotyped SARS-CoV-2 virus. +++, IC50 < 10 ng/ml; ++, IC50 = 10–100 ng/ml; +, IC50 = 100–1000 ng/ml; −, IC50 > 1000 ng/ml.
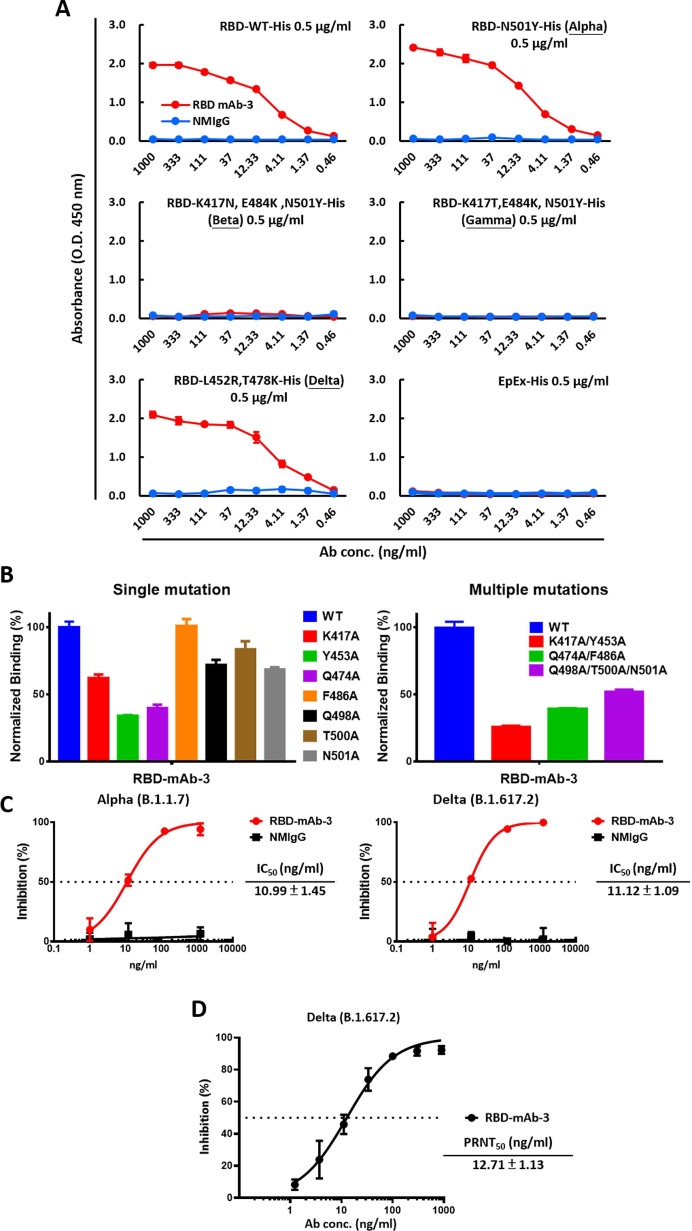
2.5. Neutralizing effect of RBD-mAb-3 against SARS-CoV-2 Alpha and Delta
As RBD-mAb-3 showed efficient neutralization of Alpha and Delta SARS-CoV-2 variant pseudoviruses, it was selected for further IgG antibody purification and validation. To detect the binding activities of purified RBD-mAb-3, ELISA was carried out. As shown in Fig. 5 A, RBD-mAb-3 had high binding potencies against RBD Alpha and Delta variants. The K417, Y453, Q474, F486, Q498, T500, and N501 residues within the RBD are known to be responsible for S protein contact with the host receptor, ACE2 (Yan et al., 2020). A cellular ELISA-based binding assay was performed to evaluate whether RBD-mAb-3 binding is affected by alanine mutations at positions K417, Y453, Q474, F486, Q498, T500 and N501 (Fig. 5B). The results showed that Y453A and Q474A mutations significantly decreased the binding of RBD-mAb-3, suggesting that these residues contribute to the RBD-mAb-3 epitope. We then further examined the neutralizing abilities of purified RBD-mAb-3 toward SARS-CoV-2 Alpha and Delta pseudoviruses. Notably, RBD-mAb-3 exhibited high neutralizing capacities for Alpha and Delta variants, with respective IC50 values of 10.99 and 11.12 ng/ml (Fig. 5C). Given the specificity and binding affinity of RBD-mAb-3 towards the Delta RBD variant, we also tested its neutralization potential in a plaque reduction neutralization test (PRNT) using authentic SARS-CoV-2 Delta variant (Fig. 5D). RBD-mAb-3 effectively neutralized viral infection with a PRNT50 value of 12.71 ng/ml. Collectively, these data indicate that RBD-mAb-3 is a potent neutralizing mAb for SARS-CoV-2 Delta variant.
Fig. 5.
RBD-mAb-3 exhibits potent binding and neutralization of SARS-CoV-2 Delta. (A) The binding of RBD-mAb-3 to different variants of SARS-CoV-2 RBDs was determined by ELISA analysis. EpEX served as a negative control. (B) Epitope mapping of RBD-mAb-3 using mutagenesis. RBD-mAb-3 binding to mutant RBDs with single or multiple alanine mutations was normalized to its binding to wild-type (WT) RBD; measurements were made with cellular ELISA. (C) Neutralization assays were performed on SARS-CoV-2 Alpha and Delta pseudoviruses with purified RBD-mAb-3. Assays were performed in triplicate; each point represents the mean ± SEM. IC50 values were calculated with GraphPad Prism software. (D) RBD-mAb-3 neutralizes SARS-CoV-2 Delta according to PRNT. The PRNT50 value was calculated with GraphPad Prism software. Assays were performed in triplicate, and points represent the mean ± SD.
3. Discussion
Throughout the SARS-CoV-2 pandemic, mAbs have gained widespread use in treatments and diagnostic tools. For example, several neutralizing mAbs have been granted emergency use authorizations (EUAs) for treatment of non-hospitalized patients with mild-to-moderate COVID-19 (Hwang et al., 2022, Taylor et al., 2021). Hybridoma technology is a highly reproducible and scalable method used to generate mAbs, which enables the production of highly specific and pure antibodies. Hybridomas are hybrid cell lines formed by fusion of a short-lived antibody-producing B cell and an immortal myeloma cell. A major advantage of this method is that it is cost-effective, as stable hybridoma cells constitutively express large amounts of one specific mAb, and the preferred cell lines can be cryopreserved for continuous mAb production. Since hybridoma technology is one of the most reliable approaches for mAb production, it has been widely applied in many contexts (Lu et al., 2020). However, there are still several limitations worth noting. First of all, preparation of traditional protein antigens is often the most critical and time-consuming step in the mAb production process, especially for mAbs against hard-to-express and -purify antigens like membrane proteins, glycoproteins or conformational epitopes. Therefore, mRNA-LNP immunization may be a more effective strategy to quickly produce mAbs against such targets. The COVID-19 pandemic has accelerated the research toward mRNA technologies (Kumar et al., 2022), and the success of mRNA-LNP vaccines suggests that the technology may be efficiently applied for the rapid development of high-quality mAbs against SARS-CoV-2 and other pathogens. In this study, we demonstrated this potential by using RBD mRNA-LNP as a novel immunogen for hybridoma-based mAb production. The main advantage of mRNA-LNP immunization is that the mRNA is easily produced by IVT and amenable to rapid manipulation of antigens. The only custom-produced material necessary for this method is the plasmid encoding an optimized target sequence. Moreover, following cellular uptake of the nucleoside-modified mRNA-LNP, abundant protein is produced with native folding. Thus, generation of mAbs using mRNA-LNP immunization can recapitulate native antigen structure. In addition, our LNPs include D-Lin-MC3-DMA, a commercially available cationic lipid, which can facilitate efficient encapsulation and delivery of RBD mRNA to cells in vitro and in vivo. Most importantly, even after antigen is produced, the process of generating hybridoma-based mAbs normally takes an average of 6 to 8 months to complete, with animal immunization being the most time-consuming step. Using the mRNA-LNP immunization strategy, we were able to successfully generate several hybridomas secreting antigen-specific mAbs while reducing the time from initial immunization to spleen harvest from 10 to 7 weeks. During the process of cell fusion and hybridoma isolation, the overall viability of cells is quite low, reducing the variety of useful antibodies that can be produced against an antigen. In addition, the human anti-mouse antibody (HAMA) response often limits the clinical applicability of murine mAbs, so an additional step of mAb humanization by CDR-grafting is usually required (Lu et al., 2020).
An alternative efficient approach to generate monoclonal antibodies from humans and immunized animals is single B cell antibody technology, which allows for the rapid isolation of potential mAbs through direct amplification of VH and VL region encoding genes from single B cells (Pedrioli and Oxenius, 2021). In order to enhance cell sorting performance in this method, fresh blood samples with an appropriate anticoagulant are required. Remarkably, this step is necessary to yield correctly paired VH and VL Fab. Based on previous studies, the rates of successful identification of VH and VL gene pairs in single-sorted B cells ranged from 40 to 60% (Fu et al., 2016, Lin et al., 2014). Such a low recovery rate requires more B cells to be sorted and analyzed, making this approach significantly more time and resource consuming. In the future, application of mRNA-LNP immunization to human antibody-producing transgenic mice (Murphy et al., 2014, Lu et al., 2020) coupled with high throughput screening of single B cells may be an even more time-efficient strategy for generating of mAbs to be used in therapeutics or diagnostics.
4. Conclusions
The majority of mAbs, including many licensed mAb drugs, were developed through hybridoma technology based on the immunization of animals with protein antigens. Despite such immunization methods having been generally successful, there still exist some limitations. The successful delivery of mRNA encoding protein of interest paved a new way for immunogen production, does not require purification of proteins. In this study, our data provide valuable insight for the design of a novel and powerful mRNA-LNP immunization method which can simplify the antigen preparation process while still eliciting good immune responses for host animals to produce high-quality mAbs.
5. Materials and methods
5.1. Production of modified IVT mRNA
The DNA template contained a T7 promoter followed by a codon optimized wild-type (Wuhan-Hu-1, GenBank YP_009724390.1) RBD sequence of SARS-CoV-2 S protein (Arg319-Phe541) flanked by the 5′ UTR, IgG kappa leader sequence and the 3′ UTR of the α-globin gene. A unique EcoRV site was included downstream of the 3′ UTR and poly(A) tail region. Prior to the IVT reaction, the plasmid was linearized using the EcoRV and purified with the NucleoSpin Gel and PCR Clean-up Kit (Macherey & Nagel Co. Düren, Germany). mRNA was synthesized by IVT according to the manufacturer’s recommendations, using HiScribe T7 (NEB, MA, USA) with co-transcriptional CleanCap® AG (Trilink, CA, USA) and N1-methyl-pseudouridine (Trilink, CA, USA). Synthesized mRNA was purified by DNase I (NEB, MA, USA) digestion followed by LiCl (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) precipitation and 70% ethanol wash. To remove dsRNA from the transcribed mRNA, selective adsorption of the dsRNA to cellulose (Sigma-Aldrich, MA, USA) was performed in an ethanol-containing buffer. Purified mRNA was kept at −80 °C until further use.
5.2. Preparation of RBD mRNA-LNP
Lipid-nanoparticle (LNP) formulations were prepared using a previously described method (Lee et al., 2022). Briefly, lipids were solubilized in ethanol containing cationic lipid (D-Lin-MC3-DMA) (MedChemExpress, NJ, USA), 1, 2-distearoyl-sn-glycero-3-phosphocholine (DSPC) (Avanti Polar Lipids, NY, USA), cholesterol (Sigma, MA, USA) and DMG-PEG 2000 (MedChemExpress, NJ, USA) at molar ratios of 50:10:38.5:1.5. The lipid mixture was combined with 25 mM sodium acetate buffer (pH 4.5) containing mRNA at a ratio of 1:3 using NanoAssemblr® IGNITE NxGen Cartridges (Precision NanoSystems Inc., BC, Canada). LNP-encapsulated mRNA samples were dialyzed against PBS (pH 7.4) at 4 °C and then concentrated using Amicon Ultra Centrifugal Filters (10 K MWCO; Millipore, Burlington, MA, USA) before being passed through 0.45-μm filters.
5.3. Characterization of RBD mRNA-LNP
The particle size distribution, polydispersity index (PDI) value and zeta potential of RBD mRNA-LNP was analyzed by Dynamic Light Scattering (DLS, Zetasizer Nano ZS, Malvern Panalytical ltd., Malvern, WR, UK). The sample was diluted 100-fold and equilibrated for 120 s at 25 °C prior to size and zeta potential measurements. The hydrodynamic diameter (z-average) and zeta potential of RBD mRNA-LNP was analyzed by Zetasizer software, version 7.11 (https://www.malvern.com). The morphology of RBD mRNA-LNP in a dry state was observed using cryogenic transmission electron microscopy (cryo-TEM, Tecnai F20, Philips, Eindhoven, the Netherlands). Briefly, the sample solution was diluted 10-fold and transferred onto 300-mesh copper grids covered with porous carbon film (HC300-Cu, PELCO) for blotting and plunging in a 100% humidity temperature-controlled chamber using a Vitroblot system (FEI). The copper grids were stored in liquid nitrogen and transferred to the electron microscope on a cryo-stage for imaging. The mRNA encapsulation efficiency (EE%) and concentration was determined with a Quant-iT RiboGreen RNA assay kit (Invitroge, Thermo Fisher Scientific, Waltham, MA, USA). The agarose gel retardation assay was performed to analyze the size and integrity of bound/unbound mRNA. The bound/unbound mRNA in LNP nanoparticles was analyzed in the presence and absence of 1% Triton X-100.
5.4. In vitro RBD expression of RBD mRNA and mRNA-LNP
RBD mRNA was transfected into HEK293T cells via Lipofectamine™ 2000 Transfection Reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA), while RBD mRNA-LNP was added directly to cells cultured in DMEM containing 10% FBS. The supernatants were collected at 48 h after transfection, and RBD expression was monitored by Western blot and sandwich ELISA. Sandwich ELISA was used to quantitatively measure the amount of RBD protein in cell culture media. Briefly, a mouse anti-RBD mAb was diluted to a concentration of 2 μg/ml in 0.1 M sodium carbonate buffer (NaHCO3/Na2CO3; pH 9.5). Then, a 96-well plate was coated with coating buffer at 4 °C overnight. The next day, the coating buffer was discarded, and the plate was blocked with PBS containing 1% bovine serum albumin (BSA) at room temperature for 1 h. The coated plates were washed three times with PBS and incubated with calibrator solutions or serial dilutions of cell culture media at RT for 1 h. After washing the plates three times with 0.1% Tween-20 in PBS (PBST0.1), anti-RBD chAb diluted in 1% BSA/PBS at 1:2000 was applied at room temperature for 1 h. The plates were washed with PBST0.1 three times, then incubated with horseradish peroxidase (HRP)-conjugated anti-human IgG (Sigma, MA, USA) secondary antibody at room temperature for 1 h. After three washes with PBST0.1, signal was generated using 3,3′5,5′-Tetramethylbenzidine (TMB) substrate (TMBW-1000–01, SURMODICS). The reaction was stopped with 3 N HCl, and absorbance was measured at 450 nm by ELISA reader (Versa Max Tunable Microplate Reader; Molecular Devices).
5.5. Mouse immunization
All procedures involving animals were performed in accordance with guidelines set by the Institutional Animal Care and Use Committee (IACUC) at Academia Sinica, Taiwan. BALB/c mice 6 to 8 weeks of age were immunized by four intramuscular injections of 10 μg RBD mRNA-LNP at 2-week intervals or four intraperitoneal injections of 50 μg recombinant RBD protein in CFA-IFA adjuvant at 3-week intervals (n = 3 per group). Serum samples were collected from each mouse before the priming injection and at one week after the first or second booster.
5.6. Generation of SARS-CoV-2 RBD-specific murine mAbs
Hybridomas secreting anti-RBD mAbs were generated and identified according to standard procedures (Köhler and Milstein, 1975) with some modifications (Chen et al., 2007, Tung et al., 2013). Briefly, the spleen of an immunized mouse was removed, and splenocytes were fused with NSI/1-Ab4–1 (NS-1) myeloma cells. The fused cells were cultured in DMEM supplemented with 15% FBS, hypoxanthine-aminopterin-thymidine (HAT) medium and hybridoma cloning factor, in 96-well tissue culture plates. Hybridoma colonies were screened by ELISA for secretion of mAbs that bound to RBD recombinant protein. Selected clones were subcloned by limiting dilutions. Final hybridoma clones were isotyped using an isotyping kit. Ascitic fluids were produced in IFA-primed BALB/c mice. Hybridoma cell lines were grown in DMEM with 10% heat-inactivated FBS. The candidate mAbs were affinity purified with protein G Sepharose 4B gels. ELISA assays were used to measure the activity and specificity of each antibody.
5.7. Antibody binding to SARS‑CoV‑2 RBD protein variants by ELISA
Recombinant His-tagged RBD proteins from SARS-CoV-2 variants were purchased as lyophilized powders from ACROBiosystems Inc. (Newark, DE, USA). The 96-well ELISA plates were coated with 0.5 μg/ml SARS-CoV-2 variant RBD-His or EpEX-His (negative control) proteins in 0.1 M sodium carbonate buffer at 4 °C overnight, followed by blocking with 1% BSA in PBS at room temperature for 2 h. After blocking, the coated plates were washed twice with PBS and stored at −20 °C until further use. The protein contents of the culture supernatants from hybridomas or antibodies were quantified by the BCA assay, and serial dilutions were performed with 1% BSA in PBS. Then, 50 μl of the hybridoma supernatant or antibody dilutions were added to individual wells, and the plate was incubated at room temperature for 1 h. The plates were washed with PBS containing 0.1% Tween-20 (PBST0.1) three times and then incubated for 1 h with Peroxidase AffiniPure Goat anti-mouse IgG (H + L) (Jackson ImmunoResearch) (1:5000 dilution). After three washes with PBST0.1, TMB signal was generated and detected.
5.8. Pseudovirus neutralization assay
SARS-CoV-2 pseudotyped lentiviruses expressing full-length S proteins and HEK293T cells overexpressing human ACE2 (HEK293T/hACE2) were provided by the National RNAi Core Facility (Academia Sinica, Taiwan). The pseudovirus neutralization assays were performed using serial dilutions of RBD-mAbs pre-incubated with 1000 TU SARS-CoV-2 pseudovirus in 1% FBS DMEM for 1 h at 37 °C. The dilutions were incubated for 24 h at 37 °C in 96-well white plates (SPL Life Science), which had been pre-seeded with HEK293T/hACE2 cells at a density of 1 × 104 cells per well. The pseudovirus-containing culture medium was then replaced with 10% FBS DMEM for an additional 48 h. Next, ONE-Glo luciferase reagent (Promega) was added to each well for a 3 min incubation at 37 °C. Luminescence was measured using a microplate spectrophotometer (Molecular Devices) to determine pseudovirus infection efficacy. The half maximal inhibitory concentration (IC50) was calculated by nonlinear regression using Prism software version 8.1.0 (GraphPad Software Inc.).
5.9. Cellular ELISA
HEK293T cells were transiently transfected with wild-type or mutant RBD plasmids in 6-well plates for 16 h. Then, the cells were seeded into 96-well plates at a density of 3 × 104 cells per well and incubated for one day. The cells were fixed in 4% paraformaldehyde in PBS for 15 min at room temperature, and then the cell membranes were permeabilized using 0.1% Triton X-100 at room temperature for 10 min. The plates were blocked using 5% milk, and 100 ng/ml RBD-mAb was added to each well for 1 h at room temperature. Next, the horseradish peroxidase (HRP)-conjugated anti-mouse antibody (1:2000) was added for 1 h at room temperature. The binding capacities of each RBD-mAb to the RBD mutants were examined by ELISA.
5.10. Plaque reduction neutralization test (PRNT)
RBD-mAb-3 was serially diluted in PBS and pre-incubated with 100 plaque-forming units (PFU) of SARS-CoV-2 for 1 h at 37 °C. The mixtures were added to pre-seeded Vero E6 cells for 1 h at 37 °C. The virus-containing culture medium was then removed and replaced with DMEM containing 2% FBS and 1% methyl-cellulose for an additional 4 days incubation. The cells were fixed with 10% formaldehyde overnight and stained with 0.5% crystal violet for 20 min. The plates were then washed with tap water, and plaque numbers formed were counted for each dilution. Virus without RBD-mAb-3 served as a control. Each experiment was performed in triplicate. Plaque reduction was calculated as: Inhibition percentage = 100 × [1 – (plaque number with mAb/plaque number without mAb)]. The 50% plaque reduction (PRNT50) value was calculated with Prism software. The SARS-CoV-2 Delta variant (hCoV-19/Taiwan/1144/2021) was obtained from Taiwan Centers for Disease Control (CDC). The PRNT assay was performed in a BSL-3 facility in the Institute of Biomedical Sciences, Academia Sinica.
Funding
This research was funded by Academia Sinica, Key and Novel Therapeutics Development Program for Major Diseases (AS-KPQ-111-KNT) and the Emerging Infectious and Major Disease Research Program (AS-KPQ-111-EIMD) to HCW.
Authors contributions
FFH designed and performed experiments, and wrote the manuscript. FFH, WYC and HTL immunized the mice and generated mouse mAbs. KHL performed in vitro neutralization by pseudoviruses. WYC performed neutralizing Ab expression. FFH, MK, MHT and CMC synthesized mRNA-LNP. HCW conceived the experiments, obtained funding, wrote and revised the paper, and provided overall direction for the study.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We are indebted to the Human Therapeutic Ab R&D Core facility of BioTReC, Academia Sinica for assisting Ab production and analysis. We also thank for the National RNAi Core Facility at Academia Sinica for providing SARS-CoV-2 variant pseudoviruses.
Footnotes
Supplementary material to this article can be found online at https://doi.org/10.1016/j.ijpharm.2022.122256.
Appendix A. Supplementary material
The following are the Supplementary material to this article:
Supplementary Fig. 1.
Supplementary Fig. 2.
Supplementary Fig. 3.
Data availability
Data will be made available on request.
References
- Baptista B., Carapito R., Laroui N., Pichon C., Sousa F. mRNA, a revolution in biomedicine. Pharmaceutics. 2021;13:2090. doi: 10.3390/pharmaceutics13122090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman A.P., Tang X., Lee J.R., Chida A., Mercer K., Wharton R.E., Kainulainen M., Harcourt J.L., Martines R.B., Schroeder M., Zhao L., Bryksin A., Zhou B., Bergeron E., Bollweg B.C., Tamin A., Thornburg N., Wentworth D.E., Petway D., Bagarozzi D.A., Jr, Finn M.G., Goldstein J.M. Rapid development of neutralizing and diagnostic SARS-COV-2 mouse monoclonal antibodies. Sci. Rep. 2021;11:9682. doi: 10.1038/s41598-021-88809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-C., Huang H.-N., Lin C.-T., Chen Y.-F., King C.-C., Wu H.-C. Generation and characterization of monoclonal antibodies against dengue virus type 1 for epitope mapping and serological detection by epitope-based peptide antigens. Clin. Vaccine Immunol. 2007;14(4):404–411. doi: 10.1128/CVI.00249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarella P., Fazio V.M. Mouse monoclonal antibodies in biological research: strategies for high-throughput production. Biotechnol. Lett. 2008;30(8):1303–1310. doi: 10.1007/s10529-008-9706-5. [DOI] [PubMed] [Google Scholar]
- Du W., Hurdiss D.L., Drabek D., Mykytyn A.Z., Kaiser F.K., González-Hernández M., Muñoz-Santos D., Lamers M.M., van Haperen R., Li W., Drulyte I., Wang C., Sola I., Armando F., Beythien G., Ciurkiewicz M., Baumgärtner W., Guilfoyle K., Smits T., van der Lee J., van Kuppeveld F.J.M., van Amerongen G., Haagmans B.L., Enjuanes L., Osterhaus A.D.M.E., Grosveld F., Bosch B.J. An ACE2-blocking antibody confers broad neutralization and protection against Omicron and other SARS-CoV-2 variants of concern. Sci. Immunol. 2022;26:eabp9312. doi: 10.1126/sciimmunol.abp9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Zhang Z., Sheehan J., Avnir Y., Ridenour C., Sachnik T., Sun J., Hossain M.J., Chen L.M., Zhu Q., Donis R.O., Marasco W.A. A broadly neutralizing anti-influenza antibody reveals ongoing capacity of haemagglutinin-specific memory B cells to evolve. Nat. Commun. 2016;7:12780. doi: 10.1038/ncomms12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzlöhner P., Hanack K. Generation of murine monoclonal antibodies by hybridoma technology. J. Vis. Exp. 2017;119:54832. doi: 10.3791/54832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021;6(12):1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y.C., Lu R.M., Su S.C., Chiang P.Y., Ko S.H., Ke F.Y., Liang K.H., Hsieh T.Y., Wu H.C. Monoclonal antibodies for COVID-19 therapy and SARS-CoV-2 detection. J. Biomed. Sci. 2022;29:1. doi: 10.1186/s12929-021-00784-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ickenstein L.M., Garidel P. Lipid-based nanoparticle formulations for small molecules and RNA drugs. Expert Opin. Drug Deliv. 2019;16(11):1205–1226. doi: 10.1080/17425247.2019.1669558. [DOI] [PubMed] [Google Scholar]
- Jackson N.A.C., Kester K.E., Casimiro D., Gurunathan S., DeRosa F. The promise of mRNA vaccines: a biotech and industrial perspective. npj Vaccines. 2020;5:11. doi: 10.1038/s41541-020-0159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256(5517):495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Kumar A., Blum J., Thanh Le T., Havelange N., Magini D., Yoon I.-K. The mRNA vaccine development landscape for infectious diseases. Nat. Rev. Drug Discov. 2022;21(5):333–334. doi: 10.1038/d41573-022-00035-z. [DOI] [PubMed] [Google Scholar]
- Kumari M., Lu R.M., Li M.C., Huang J.L., Hsu F.F., Ko S.H., Ke F.Y., Su S.C., Liang K.H., Yuan J.P., Chiang H.L., Sun C.P., Lee I.J., Li W.S., Hsieh H.P., Tao M.H., Wu H.C. A critical overview of current progress for COVID-19: development of vaccines, antiviral drugs, and therapeutic antibodies. J. Biomed. Sci. 2022;29:68. doi: 10.1186/s12929-022-00852-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I.J., Sun C.P., Wu P.Y., Lan Y.H., Wang I.H., Liu W.C., Yuan J.P., Chang Y.W., Tseng S.C., Tsung S.I., Chou Y.C., Kumari M., Lin Y.S., Chen H.F., Chen T.Y., Lin C.C., Chiu C.W., Hsieh C.H., Chuang C.Y., Cheng C.M., Lin H.T., Chen W.Y., Hsu F.F., Hong M.H., Liao C.C., Chang C.S., Liang J.J., Ma H.H., Chiang M.T., Liao H.N., Ko H.Y., Chen L.Y., Ko Y.A., Yu P.Y., Yang T.J., Chiang P.C., Hsu S.T., Lin Y.L., Lee C.C., Wu H.C., Tao M.H. A booster dose of Delta × Omicron hybrid mRNA vaccine produced broadly neutralizing antibody against Omicron and other SARS-CoV-2 variants. J. Biomed. Sci. 2022;29:49. doi: 10.1186/s12929-022-00830-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Chiang N.Y., Chai N., Seshasayee D., Lee W.P., Balazs M., Nakamura G., Swem L.R. In vivo antigen-driven plasmablast enrichment in combination with antigen-specific cell sorting to facilitate the isolation of rare monoclonal antibodies from human B cell. Nat. Protoc. 2014;9:1563–1577. doi: 10.1038/nprot.2014.104. [DOI] [PubMed] [Google Scholar]
- Liu S., Wang S., Lu S. DNA immunization as a technology platform for monoclonal antibody induction. Emerg. Microbes Infect. 2016;5(1):e33. doi: 10.1038/emi.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R.M., Hwang Y.C., Liu I.J., Lee C.C., Tsai H.Z., Li H.J., Wu H.C. Development of therapeutic antibodies for the treatment of diseases. J. Biomed. Sci. 2020;27:1. doi: 10.1186/s12929-019-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusvarghi S., Pollett S.D., Neerukonda S.N., Wang W., Wang R., Vassell R., Epsi N.J., Fries A.C., Agan B.K., Lindholm D.A., Colombo C.J., Mody R., Ewers E.C., Lalani T., Ganesan A., Goguet E., Hollis-Perry M., Coggins S.A., Simons M.P., Katzelnick L.C., Wang G., Tribble D.R., Bentley L., Eakin A.E., Broder C.C., Erlandson K.J., Laing E.D., Burgess T.H., Mitre E., Weiss C.D. SARS-CoV-2 BA.1 variant is neutralized by vaccine booster-elicited serum but evades most convalescent serum and therapeutic antibodies. Sci. Transl. Med. 2022;14:eabn8543. doi: 10.1126/scitranslmed.abn8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruggi G., Zhang C., Li J., Ulmer J.B., Yu D. mRNA as a transformative technology for vaccine development to control infectious diseases. Mol. Ther. 2019;27(4):757–772. doi: 10.1016/j.ymthe.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy A.J., Macdonald L.E., Stevens S., Karow M., Dore A.T., Pobursky K., Huang T.T., Poueymirou W.T., Esau L., Meola M., Mikulka W., Krueger P., Fairhurst J., Valenzuela D.M., Papadopoulos N., Yancopoulos G.D. Mice with megabase humanization of their immunoglobulin genes generate antibodies as efficiently as normal mice. Proc. Natl. Acad. Sci. U. S. A. 2014;111(14):5153–5158. doi: 10.1073/pnas.1324022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17(4):261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr C.J.C., Wada S., Kotake K., Kameda S., Matsuura S., Sakashita S., Park S., Sugiyama H., Kuang Y.i., Saito H. N1-Methylpseudouridine substitution enhances the performance of synthetic mRNA switches in cells. Nucleic Acids Res. 2020;48(6):e35. doi: 10.1093/nar/gkaa070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parray H.A., Shukla S., Samal S., Shrivastava T., Ahmed S., Sharma C., Kumar R. Hybridoma technology a versatile method for isolation of monoclonal antibodies, its applicability across species, limitations, advancement and future perspectives. Int. Immunopharmacol. 2020;85:106639. doi: 10.1016/j.intimp.2020.106639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrioli A., Oxenius A. Single B cell technologies for monoclonal antibody discovery. Trends Immunol. 2021;42(12):1143–1158. doi: 10.1016/j.it.2021.10.008. [DOI] [PubMed] [Google Scholar]
- Sun P., Lu X., Xu C., Sun W., Pan B. Understanding of COVID-19 based on current evidence. J. Med. Virol. 2020;92:548–551. doi: 10.1002/jmv.25722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda H., Ogasawara T., Ozawa T., Muraguchi A., Jih P.J., Morishita R., Uchigashima M., Watanabe M., Fujimoto T., Iwasaki T., Endo Y., Sawasaki T. Production of monoclonal antibodies against GPCR using cell-free synthesized GPCR antigen and biotinylated liposome-based interaction assay. Sci. Rep. 2015;5:11333. doi: 10.1038/srep11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P.C., Adams A.C., Hufford M.M., de la Torre I., Winthrop K., Gottlieb R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021;21(6):382–393. doi: 10.1038/s41577-021-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung K.H., Lin C.W., Kuo C.C., Li L.T., Kuo Y.H., Lin C.W., Wu H.C. CHC promotes tumor growth and angiogenesis through regulation of HIF-1alpha and VEGF signaling. Cancer Lett. 2013;331:58–67. doi: 10.1016/j.canlet.2012.12.001. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang Z., Luo J., Han X., Wei Y., Wei X. mRNA vaccine: a potential therapeutic strategy. Mol. Cancer. 2021;20:33. doi: 10.1186/s12943-021-01311-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L.u., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in china, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X., Mattern M.R., Tan R., Li S., Hall J., Sterner D.E., Shoo J., Tran H., Lim P., Sarafianos S.G., Kazi L., Navas-Martin S., Weiss S.R., Butt T.R. Expression and purification of SARS coronavirus proteins using SUMO-fusions. Protein Expr. Purif. 2005;42(1):100–110. doi: 10.1016/j.pep.2005.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.