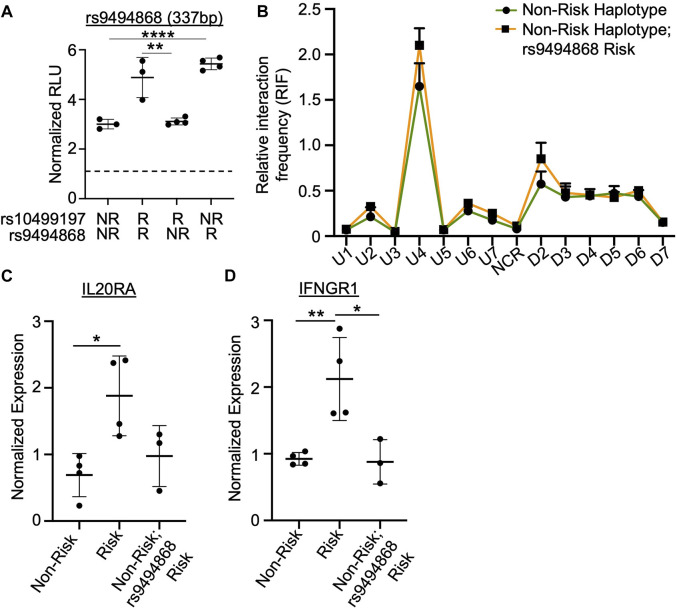
FIGURE 5.
SNP rs9494868 alters the in vitro binding affinities and enhancer activity, but not chromatin-chromatin looping to this enhancer. (A) Dual luciferase assay using EBV B cells transiently transfected with empty pGL4.23 or pGL4.23 containing the 337bp construct with the indicated non-risk (T) or risk (G) allele of rs10499197 and non-risk (T) or risk (G) allele of rs9494868. Relative luciferase units were calculated as a normalized ratio of Firefly to Renilla; Student’s t-test, n > 3; ****p < 0.001 (Figure Abbreviations: V, pGL4.23 vector-only; NR, pGL4.23 with non-risk allele; R, pGL4.23 with risk allele). (B) 3C-qPCR was performed in EBV B cells homozygous for the SLE TNFAIP3 non-risk haplotype and either the non-risk (T) or risk (G) allele of rs9494868 using primers shown in Figure 3A. Relative interaction frequency was calculated by normalizing the interaction frequencies to the interaction frequency of the random ligation control and plotted as mean ± SEM, n = 3; Student’s t-test, *p < 0.05. Interactions were considered positive if the relative interaction frequency was greater than the interactions between the anchor and the negative control region of the locus ligation control. (C,D) Expression of IL20RA (C) and IFNGR1 (D) were measured in EBV B cells for the SLE TNFAIP3 non-risk haplotype, SLE TNFAIP3 risk haplotype, or SLE TNFAIP3 non-risk haplotype and the risk allele (G) of rs9494868 using qRT-PCR. Expression was normalized to GAPDH; Student’s t-test, n > 3; *p < 0.05; **p < 0.01.