Fig. 1.
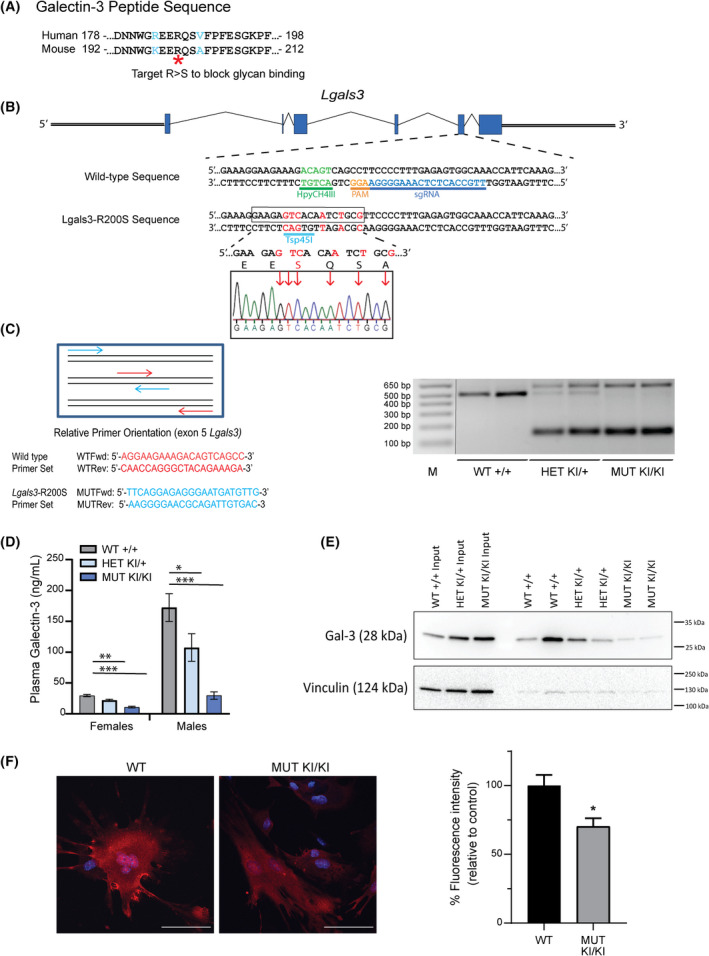
Generation of Lgals3‐R200S knock‐in allele by CRISPR/Cas9. (A) Comparison of human and mouse Gal‐3 amino acid sequence and identification of an essential arginine to mutate to generate the glycan‐binding deficient Gal‐3. (B) The target sequence of the Lgals3 gene showing PAM and sgRNA binding site. Red letters indicate nucleotides to be mutated following homologous recombination of the single‐stranded oligonucleotide (ssODN) template. Sanger sequencing confirmation of incorporation of mutations. (C) PCR mediated identification of mice with wild‐type and Lgals3‐R200S alleles. M, molecular weight ladder; MUT, mutant; WT, wild‐type. Expected amplified product sizes are 483 bp for wild‐type and 131 and 586 bp for the R200S knock‐in allele. (D) Plasma serum levels of Gal‐3 in 36‐week‐old WT (+/+) female n = 10 and male n = 12, HET (KI/+) female n = 15 and male n = 11, and MUT (KI/KI) female n = 15 and male n = 13 animals. The samples were run in duplicate and the assay was run once. Values are expressed as mean ± SEM (n = 9–15). Dunnett's post hoc analysis adjusted P‐values compared to wild‐type; bold values highlight *P < 0.05, **P < 0.01, ***P < 0.001. (E) Immunoblot of Gal‐3 and vinculin on the cell surface of mouse embryonic cells isolated from WT (+/+), HET (KI/+), and MUT (KI/KI) embryos. Two biological replicates were evaluated per genotype and the assay was run once. (F) Gal‐3 immunocytochemistry of permeabilized MEFs from Lgals3‐R200SKI/KI and WT mice. Gal‐3 is present throughout the cytosol and nucleus in both conditions. Gal‐3 protein levels are overall reduced by approximately 30%. Scale bar = 100 μm. One biological sample per genotype and five technical slide replicates per genotype were evaluated, with a total of n = 31 WT MEF cells and n = 40 Lgals3‐R200S mutant MEF cells. Quantification of Gal‐3 staining intensity is indicated on the right, data is mean ± SEM. Asterisks indicate a significant difference, *P < 0.05.
