Abstract
Frozen sections provide a basis for rapid intraoperative diagnosis that can guide surgery, but the diagnoses often challenge pathologists. Here we propose a rule-based system to differentiate thyroid nodules from intraoperative frozen sections using deep learning techniques. The proposed system consists of three components: (1) automatically locating tissue regions in the whole slide images (WSIs), (2) splitting located tissue regions into patches and classifying each patch into predefined categories using convolutional neural networks (CNN), and (3) integrating predictions of all patches to form the final diagnosis with a rule-based system. To be specific, we fine-tune the InceptionV3 model for thyroid patch classification by replacing the last fully connected layer with three outputs representing the patch's probabilities of being benign, uncertain, or malignant. Moreover, we design a rule-based protocol to integrate patches’ predictions to form the final diagnosis, which provides interpretability for the proposed system. On 259 testing slides, the system correctly predicts 95.3% (61/64) of benign nodules and 96.7% (148/153) of malignant nodules, and classify 16.2% (42/259) slides as uncertain, including 19 benign and 16 malignant slides, which are a sufficiently small number to be manually examined by pathologists or fully processed through permanent sections. Besides, the system allows the localization of suspicious regions along with the diagnosis. A typical whole slide image, with 80, 000 × 60, 000 pixels, can be diagnosed within 1 min, thus satisfying the time requirement for intraoperative diagnosis. To the best of our knowledge, this is the first study to apply deep learning to diagnose thyroid nodules from intraoperative frozen sections. The code is released at https://github.com/PingjunChen/ThyroidRule.
Keywords: Thyroid nodule, Frozen section, Whole slide image, Deep learning, Rule-based protocol
1. Introduction
Intraoperative frozen sections are very useful for rapid pathology-based diagnosis that can guide further surgical decisions [1], but making diagnoses from frozen sections is very challenging even for experienced pathologists: the quality of frozen sections is lower compared to formalin fixed paraffin embedded tissue, the samples may contain artifacts, and they must be diagnosed within 20 min of receipt. Well-trained pathologists who are qualified to make diagnoses from frozen sections are less common [2], and a net gap of 5700 pathologists is expected by 2030 in the USA alone [3]. The shortage of pathologists is particularly severe in developing countries, such as sub-Saharan Africa [4,5]. China has a similar number of pathologists as the USA (approximately 20,000) but three times the population [6,7].
One potential solution to the challenges of making rapid, accurate diagnoses for frozen sections is a type of artificial intelligence called deep learning, which has found substantial applications and success within medicine [8-11] as well as outside [12-14]. Following its success in image recognition [15], semantic segmentation [16], object detection [17], etc., deep learning is rapidly introduced into the medical domain. For example, deep neural networks can classify skin lesions with accuracy similar to that of dermatologists [18], and they can detect diabetic retinopathy and macular edema in retinal fundus photographs with a sensitivity of 97.5% and specificity of 93.4% [19]. Moreover, deep learning has been applied to digital pathology and has shown promising results in multiple applications. As early as 2014, Wang et al. start to use convolutional neural networks (CNN) to extract features to detect mitoses to assist breast cancer grading [20]. They combine the handcrafted features with CNN features to maximize the detection performance. In 2015, Ertosun et al. propose an ensemble of CNNs for automated gliomas grading [21] and achieve a classification accuracy of 96% for the task of GBM vs. LGG classification. Since 2016, deep learning has been widely used and achieved optimal performance in many tasks of digital pathology, such as detection of invasive breast cancer [22] and lymph node metastases [23], epithelial and stromal regions segmentation and classification [24], tumor content and cellularity of prostate cancer estimation [25], nucleus/cell detection, segmentation, and retrieval [26-30], etc.
In this study, we investigate for the first time whether deep learning can diagnose thyroid nodules from intraoperative frozen sections. Thyroid nodules are one of the most common specimens requiring intraoperative consultations in current clinical practice. However, the sensitivity for diagnosing thyroid nodules from frozen sections is only around 75% [31,32]. Nevertheless, frozen sections are needed to confirm malignancy suspected from fine-needle aspiration cytology, and such sections should always be prepared in the case of supracentimetric isolated nodules, to determine the surgical strategy [33]. We reason that diagnosing thyroid nodules could be a feasible task for deep learning because analysis of frozen sections can be reduced to a three-category classification problem (benign, uncertain, or malignant) based on the guidance requirements for the following surgery decisions. Also, the most frequent thyroid malignancy, papillary thyroid carcinoma (PTC), shows distinct characteristic patterns [34,35].
Because digitized frozen section slides have very high resolution, which can be up to size 100,000 × 100,000 pixels, the computation cost would be prohibitive if directly dealing with them. Whereas large scale down-sampling would seriously lose the details, resulting in the slide to be blurry to the extent that cannot be diagnosed. Splitting the WSI into patches to conduct analysis and then fusing patches’ prediction is the most common solution for histopathology WSI analysis [36-39]. In the study of Hou et al. [36], they formulate a novel Expectation-Maximization (EM) based method to automatically locates discriminative patches. In their approach, patches labels are iterative inferred and refined, thus is free from region annotation. However, the overall computation cost would be expensive because of the iterative refinement procedure. Along with Hou's study, Wang et al. [40] propose a weakly supervised approach to maximize the use of available WSI-level labels. Furthermore, they explore context-aware block selection and feature aggregation strategies to generate globally holistic WSI descriptor. But the performance of patch-based classifier is not satisfactory under the weakly supervised manner. Maximilian et al. [41] formulate the whole slide image diagnosis as a multiple instance learning (MIL) problem and propose to learn the Bernoulli distribution of the patch label where the patch label probability is fully parameterized by neural networks. This study is theoretically graceful but it basically does not consider the diagnosis preference of certain categories in practical diagnosis. Wei et al. [38] adopt a sliding window approach to generate small patches on the whole slide, and classify each patch with a residual neural network. They use a heuristic manner to determine predominant and minor histologic patterns for the whole slide. This method is very straightforward and the label of different regions can be obtained during the inference of slide diagnosis. In addition, the heuristic manner can be robust to artifacts as well as single-patch classifications.
In this study, we take the applicability and interpretability of the proposed system as our main motivation. With this motivation, we first propose a tissue localization algorithm to segment tissue regions in the slide, thus reducing the overall slide diagnosis time cost as well as the interference of irrelevant regions. Then we split located tissue regions into patches and separately predict the category of each patch. The final diagnosis of thyroid nodules is performed as integration of patch predictions using a rule-based protocol. The proposed system can not only diagnose the category of thyroid lesion, but it also can locate suspicious lesions, which pathologists can manually examine as needed. Furthermore, the rule-based protocol allows for the interpretability of the proposed system, making the diagnosis to be more trustworthy. An overview of the proposed approach is shown in Fig. 1. The main contributions of this paper can be summarized as follows:
Fig. 1.
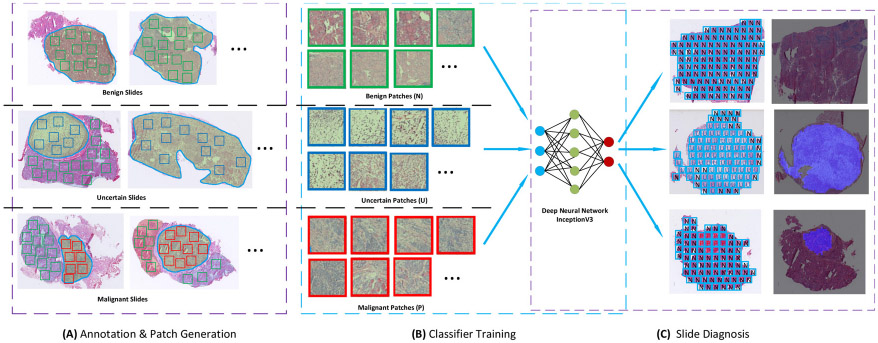
The pipeline of training and testing the model for diagnosis of thyroid nodules from frozen sections. Module (A) shows how patches of three categories are cropped from annotated slides. Uncertain patches and malignant patches are cropped from uncertain and malignant slides, respectively. Benign patches are cropped from benign regions in each of the three slide types. All cropped patches are used to fine-tune the deep learning model in module (B). In the testing stage, the trained model is applied to all patches inside localized tissues in testing slides. All patch predictions are integrated to form the final diagnosis according to a rule-based protocol. Note: “N” stands for negative, namely benign patch; “U” stands for uncertain patch; and “P” stands for positive, namely malignant patch.
We develop a rule-based diagnostic system to differentiate thyroid nodules from intraoperative frozen sections based on deep learning techniques.
Tissue localization is applied first in the whole slide diagnosis to locate thyroid tissue regions for the reduction of the overall time cost.
The rule-based system considers the conservative diagnosis manner of the practical thyroid frozen section diagnosis, thus to be a more safe system.
The automated diagnostic system obtains a precision of benign and malignant of thyroid nodules with 95.3% and 96.7% as well as 100% sensitivity of uncertain category.
2. Slides collection and categorization
We collect thyroid frozen sections and their permanent slides from Peking Union Medical College Hospital (Beijing, China) with a cohort of patients having suspected thyroid nodules admitted between January 2017 and October 2017. The hospital's Ethics Review Board approves the study. All collected slides are de-identified before the analysis. Collected frozen sections are independently reviewed by two certified pathologists, who then resolve disagreements through discussion. Permanent sections are considered as the gold standard, and used to assist the definite diagnosis of frozen sections.
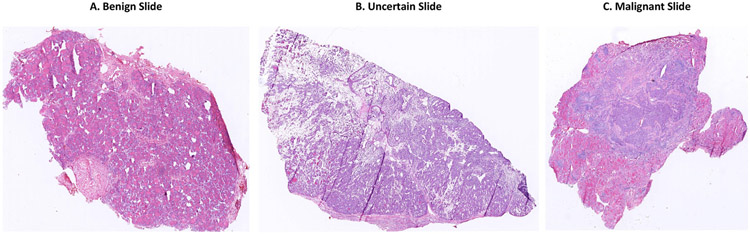
Thyroid frozen sections are categorized as benign, uncertain, or malignant, based on the requirements for developing surgery plans. Fig. 2 shows the morphology of typical thyroid slides of three categories.
Fig. 2.
Frozen section examples of (A) benign, (B) uncertain, and (C) malignant thyroid nodules.
Table 1 presents the subtypes and surgery plan for each diagnosis. Benign lesions include normal thyroid tissue, nodular hyperplasia, multinodular goiter, lymphocytic thyroiditis, Hashimoto thyroiditis, and granulomatous thyroiditis. The patient who is diagnosed as having benign lesions do not need additional surgery. Malignant lesions include papillary thyroid carcinoma, widely invasive follicular carcinoma, medullary carcinoma, poorly differentiated thyroid carcinoma, and thyroid lymphoma. Those diagnosed with malignancy need to conduct further resection, such as lobectomy or total thyroidectomy, with or without lymph node sampling or cervical lymph node dissection. Lesions that are difficult to diagnose or whose diagnosis requires sufficient sampling of the nodular capsule (follicular adenoma, minimally invasive follicular carcinoma) are defined as uncertain. Patients diagnosed as having uncertain lesions are subjected to further tissue sampling, and permanent sections are prepared and used to make a definitive diagnosis.
Table 1.
Summary of thyroid frozen sections’ diagnosis categories, all the subtypes included, and their corresponding surgery plan for each category.
| Diagnosis | Subtypes included | Surgery plan |
|---|---|---|
| Benign | Normal thyroids, nodular hyperplasia, multinodular goiter, lymphocytic thyroiditis, Hashimoto thyroiditis, granulomatous thyroiditis | Lobectomy for thyroid nodules, no more surgery |
| Malignant | PTC, medullary carcinoma, poorly differentiated thyroid carcinoma, thyroid lymphoma | Further resection, total thyroidectomy, possible lymph node sampling, or neck dissection |
| Uncertain | Follicular adenoma, follicular carcinoma, follicular PTC, adenomatous nodules | Hemithyroidectomy, deferring to permanent sections |
Note: PTC, papillary thyroid carcinoma.
3. Methods
In this section, we first describe the patch preparation and classifier selection for the patch classification model training. Then we present the three steps involved in the proposed thyroid frozen section diagnosis system in detail.
3.1. Patch classification
3.1.1. Region annotation and patch cropping
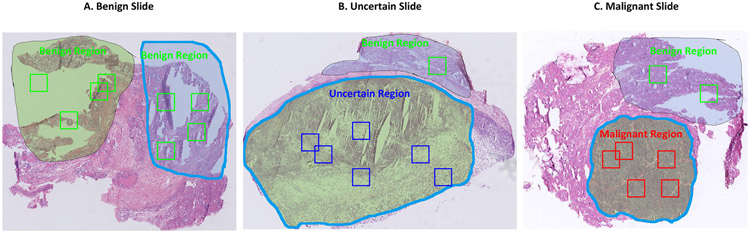
We employ supervised learning to train thyroid patch classification model, which requires sufficient annotation data. To reduce the annotation burden, we adopt the region of interests (ROI) based manner to annotate three kinds of regions. Two pathologists label all the region annotations. One pathologist first annotates the ROIs as well as their categories on all the training and validation slides. Then another pathologist checks and refines the annotations. Based on these annotations, patch images are randomly cropped from them and used as samples for deep neural network training. The label of each cropped patch is set as the same with the cropped ROI.
Fig. 3 shows annotation examples for three slides, where the region demarcated with the mask is the annotated ROI. All regions on benign slides are considered as benign, and regions from these slides are randomly selected as benign regions. We design the annotated uncertain regions on uncertain slides to include some pixels beyond the uncertain borders, while annotated malignant regions on malignant slides to be conservative to include regions that are highly likely to be malignant. Benign regions on uncertain and malignant slides are also annotated and used in training to increase the diversity of benign patches. To note that some background (non-tissue) regions on frozen sections of all three categories are also annotated as benign regions to train the patch model to avoid classifying background patches as uncertain or malignant.
Fig. 3.
Image patch cropping from annotated frozen sections. Contours are annotated and validated by certified pathologists. All regions in the benign slide are considered as benign. A few white background regions are specifically set as benign regions. Benign regions can also exist on uncertain and malignant slides. Benign, uncertain, and malignant patches are randomly cropped from these annotated regions with corresponding categories. The number of cropped patches is set to be proportional to the area of the annotated region.
We set the size of the cropped patch as 2392 × 2392 pixels on level 0 slide image, corresponding to 0.598 × 0.598 mm2 based on a pixel scale of 0.25 μm at scanning magnification of × 40. We randomly select the center coordinates for each patch and crop the patch based on the center coordinates. We only retain the patches in which at least 75% of pixels lying inside the annotated regions.
3.1.2. Patch classification model
Currently, there are several popular CNN classification architectures along the deep learning development, including VGG [42], Inception [43], ResNet [13], etc. We choose the InceptionV3 as the patch classification model for the diagnosis of thyroid nodules. The main reason to choose InceptionV3 is because of its wide adoption in medical imaging applications, such as skin cancer classification [18] and diabetic retinopathy detection [19], and its demonstrated superior performance. Based on the three-category classification for the thyroid nodule diagnosis, we replace the last fully connected layer of InceptionV3 from 1000 to 3 outputs, to represent benign, uncertain, and malignant, respectively.
Although we can generate more than 120,000 patches for classifier training, we still train the thyroid patch model via transfer learning from a pre-trained model on the ImageNet recognition task. With a pre-trained model for initialization, the patch classifier can obtain better performance, which will be shown in the experiment part.
3.2. Tissue localization
With the patch-based model, we can predict the category of all the patches inside the slide. However, before the patch-wise prediction, we need to locate the real tissue regions in the testing slide to avoid background (non-tissue) regions. As background regions not only increase the computational time but also affect the WSI diagnosis [44].
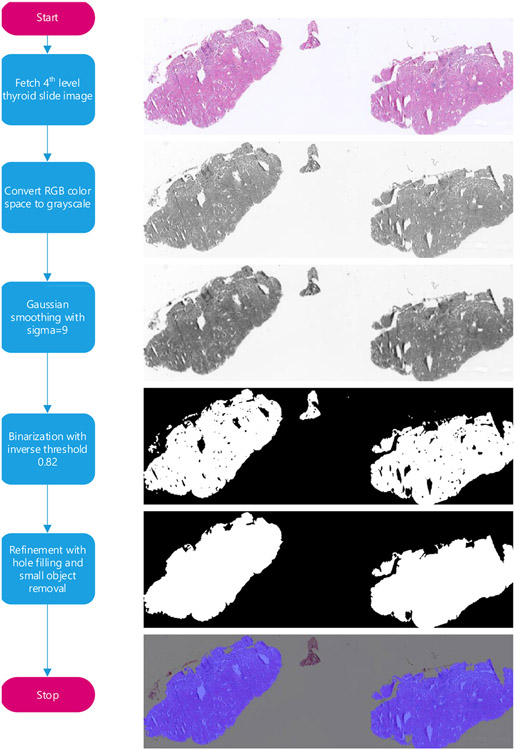
We combine a series of image processing techniques to locate tissue regions by taking advantage of the fact that the intensity value of the tissue regions is lower than the background. We perform tissue localization on the 4th level slide image, which has a width and height only 1/16 of the original WSI, which can significantly reduce the time cost for tissue localization. Firstly, the slide image is loaded from the 4th level of the whole slide image, then we convert the low-level image from RGB color space to grayscale. Gaussian filtering with a kernel size of 9 is applied to smooth the gray image. Then we inversely binarize the smoothed gray image using a preset threshold value of 0.82. Next, we apply hole filling and small object removal to refine the binary image. The refined foreground regions are considered as real tissues. Fig. 4 shows the tissue localization pipeline with intermediate results generated on each step.
Fig. 4.
Thyroid tissue localization pipeline. Tissue localization is applied at the 4th level of the whole slide image to eliminate background regions that are irrelevant to slide diagnosis, thereby speeding up the automatic diagnosis process. Color space conversion, image smoothing, inverse binarization, and binary image refinement are successively applied to generate the thyroid tissue mask.
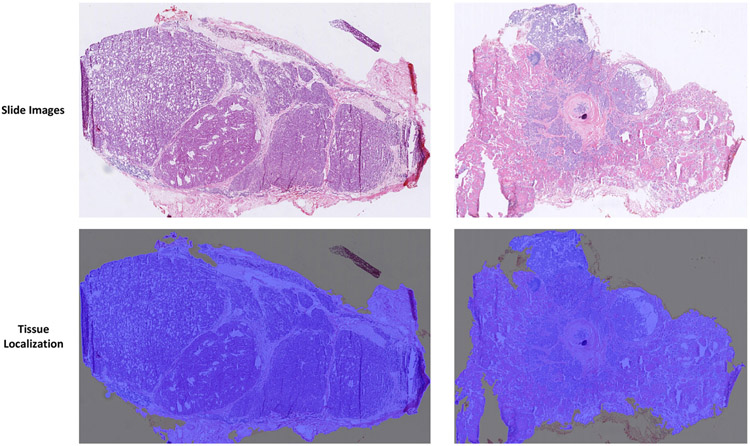
Fig. 5 shows two examples of tissue localization, in which the blue covered regions are the localized tissue regions. Parts of boundary regions can be added or missed, but this should have negligible effects on the thyroid nodule diagnosis.
Fig. 5.
Thyroid whole slide image tissue localization examples. The first row shows two sample whole slide images. The second row shows tissue localization results (marked in blue).
3.3. Patch splitting and prediction
After tissue localization, we can easily identify the tissue contours and their circumscribed rectangles. To keep the split patches with the same size as the training patches for the InceptionV3 model, we partition patches with the size of 2392 × 2392 pixels from the circumscribed rectangles in a grid-by-grid manner. To speed up the diagnosis, we set no overlap between adjacent patches in the patch splitting process. As the circumscribed rectangle covers more regions than the real tissue, there exists some patches outside or intersected with the localized contours. We keep those patches with at least 75% of pixels inside the localized tissue region for thyroid slide diagnosis.
With all the split patches from the localized tissue regions, the fine-tuned InceptionV3 model is used to make predictions on them. Each patch's probabilities belonging to the three categories are inferred and used for the subsequent patch fusion.
3.4. Fusion of patch predictions
Different from natural image classification, categories cannot always be treated equally in the medical diagnosis task. Taking the diagnosis of thyroid nodules as an example, predicting malignancy to be uncertain can cause less harm than the reverse, as the thyroid resection is irreversible. Therefore, pathologists tend to make a conservative diagnosis when dealing with a complicated case. Based on this prior knowledge, we fuse the patch predictions based on following two criteria: (1) the patch is predicted as malignant only when it is highly likely to be malignant; (2) the slide is predicted as malignant only when the predicted malignant region is larger than a preset threshold. For criterion 1, we predict the patch to be malignant only when its predicted malignant probability is higher than 0.96. The patch with predicted benign probability higher than 0.40 and the value is the biggest among the three categories will be classified as benign. The rest patches are considered as uncertain.
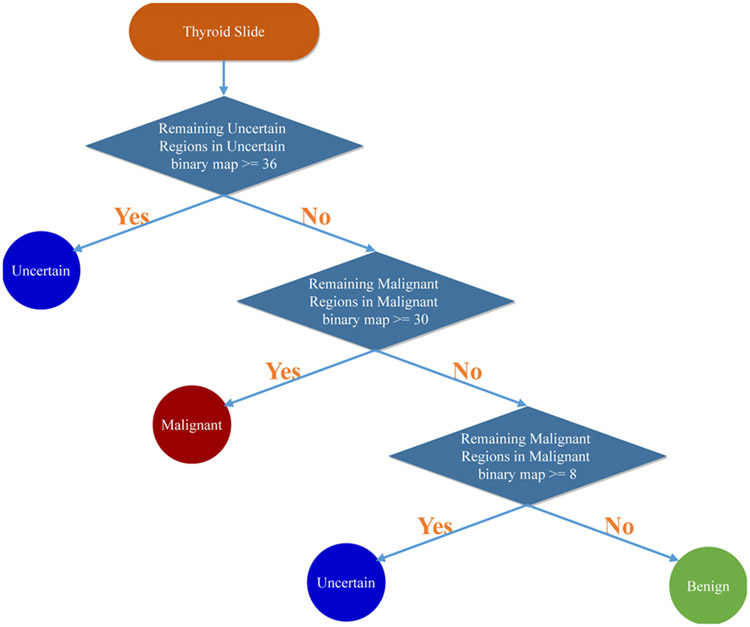
With all patches’ predicted categories, we propose a rule-based protocol to fuse all the predictions, which is mainly based on criterion 2. After classifying all the patches in the WSI, three binary maps are generated to represent benign, uncertain, and malignant, respectively. We apply hole filling to uncertain and malignant binary maps as post-processing. When the uncertain binary map contains more than 36 connected uncertain patches, the slide is diagnosed as uncertain without considering other binary maps. Next, if the malignant binary map contains more than 30 connected malignant patches, the slide will be considered as malignant. However, if the number of connected malignant patches is between 8 and 30, the slide is also diagnosed as uncertain to follow the conservative diagnosis principle. The rest slides are diagnosed as benign. This diagnosis rule is formulated as follows:
| (1) |
where Nu stands for the maximum connected number of uncertain patches in the uncertain binary map and Np stands for the maximum connected number of malignant patches in the malignant binary map. Fig. 6 shows the proposed rule-based protocol for thyroid slide diagnosis.
Fig. 6.
The proposed rule-based protocol for the diagnosis of thyroid frozen sections based on patch classification results.
All the parameters, including the probability cut-offs and patch number thresholds of different categories, are set mainly based on two factors. Firstly, we try to conform to the conservative diagnosis manner (prefer to diagnose malignant slides as benign or uncertain to avoid the non-reversible resection) adopted by pathologists used in practical diagnosis. Secondly, with the guidance of conservative diagnosis as the basis, we take advantage of the validation thyroid dataset to determine the values of these parameters based on the performance on the validation dataset. Based on this protocol, malignant and benign slides would tend to be classified as uncertain, which is demonstrated in the slide-level performance part. The main advantage of the proposed fusion method is that it can provide interpretability for the slide diagnosis. The user can know why the diagnosis is made and where those uncertain or malignant regions are located, which is very important for medical applications and makes the diagnosis to be trustworthy. Fig. 7 shows the entire thyroid frozen section diagnosis flow chart.
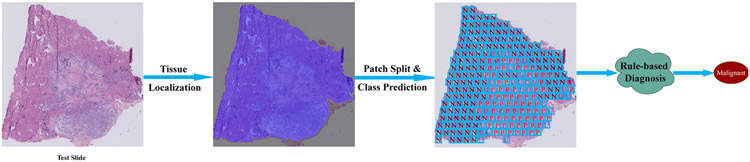
Fig. 7.
The flow chart of the proposed rule-based diagnosis system for the thyroid frozen section. The main components in the system contains tissue region localization, patch splitting and category prediction based on CNN, and the rule-based slide diagnosis protocol.
4. Experimental results
4.1. Dataset
We collect two batches of thyroid frozen sections for the system training and evaluation. The first collection contains 349 thyroid frozen sections (benign, 117; uncertain, 50; malignant, 182), is used for model construction, which is randomly divided into training and validation sets in a 4:1 ratio. Nearly all these sections came from a consecutive series of patients, except 42 slides classified as uncertain that are specifically chosen to help reduce the imbalance among the three categories. The second collection contains 259 frozen sections (benign, 85; uncertain, 7; malignant, 167), and is used for the evaluation of the proposed system.
Based on the patch cropping method, 67,031 benign, 25,843 uncertain, and 32,020 malignant patches are cropped from the training slides for model training. A further 12,487 benign, 5150 uncertain, and 7038 malignant patches are cropped from validation slides and used for classification model evaluation and selection.
4.2. Implementation details
For the patch classifier, we also train VGG16BN and ResNet50 to compare with InceptionV3 in both fine-tuning and training from scratch manner. The RMSprop [45] is used for the optimization of model parameters. The initial learning rate is set as 0.01 for training from scratch and 0.001 for fine-tuning, and it decays every two epochs with a ratio of 0.8 for both manners. Image augmentation techniques, including rotation, flipping, and color jittering, are used to improve the model's generalization ability. We train the network with a batch size of 32 and a weight decay of 0.0005 for 20 epochs. We evaluate the trained model after each epoch on all cropped validation patches. All three models with two different initialization manners are separately trained five times to compare their performance. The model with the best accuracy on validation patches is selected for the diagnosis of thyroid nodules. All the implementation are based on Python3.6, and open-soured in https://github.com/PingjunChen/ThyroidRule. We take advantage of openslide-python and scikit-image for whole slide image loading and basic image processing. We use deep learning framework PyTorch 0.4.0 to implement all the CNN models. The GeForce GTX 1080Ti GPU with 11 GB memory is used for CNN model training and evaluation. It takes about 6.4 h, 4.3 h, and 4.9 h to train the InceptionV3, VGG16BN, and ResNet50 for 20 epochs on both training from scratch and fine-tuning from the ImageNet pre-trained model.
4.3. Patch-based classification
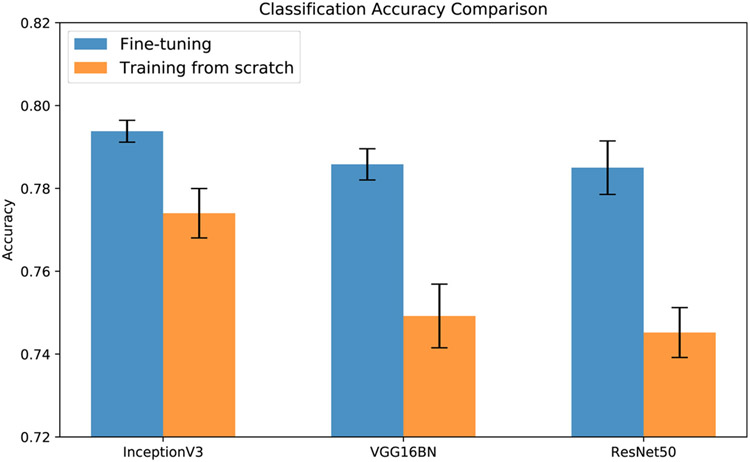
Patch classification model's performance on validation patches are shown in Fig. 8. Among the three experimented CNN models, their performance with fine-tuning training mode is very close, with InceptionV3 showing narrow lead. Whereas comparing the fine-tuning and training from scratch, it is evident that fine-tuning models achieve superior performance. These results validate the selection of InceptionV3 model via transfer learning for thyroid patch classification.
Fig. 8.
Comparison of three different classifiers, including InceptionV3, VGG16BN, and ResNet50, on thyroid patch classification via fine-tuning and training from scratch.
However, even for the best-performed InceptionV3 model, the classification accuracy on validation patches is lower than 0.80, which cannot be treated as prominent for a three-category classification task. The main reason lies in the coarse ROI-based annotation. As the ROI covers a vast region, the label of the cropped patch can be different from the category of the ROI. Thus there exists noise in the patches’ labels. Nevertheless, directly annotating patches is too time-consuming, the current ROI-based annotation is the sub-optimal solution.
4.4. Slide-level performance
The diagnosis of the testing slides is based on the best-performed InceptionV3 model and the rule-based protocol. Table 2 shows the confusion matrix of the diagnosis of thyroid nodules on 259 testing thyroid frozen sections. Of the 85 benign thyroid lesions, 61 are predicted to be benign, 19 uncertain, and 5 malignant. Among 167 malignant thyroid slides, 148 are predicted as malignant, 16 uncertain, and 3 as benign. All 7 uncertain slides are correctly predicted. Both benign and malignant categories obtain high precision, which is 95.3% (61/64) and 96.7% (148/153), respectively. The uncertain category shows a high sensitivity of 1.0. Based on the conservative diagnosis manner, benign precision, malignant precision, and the uncertain sensitivity are the most interested metrics, which all obtain prominent performance. The overall three-category classification accuracy is 83.4% (216/259). However, this underestimates the practical diagnosis accuracy, pathologists will further examine those slides classified as uncertain, and perhaps additional tissue would be prepared or fixed permanently. If we exclude those slides that are incorrectly classified as uncertain, we can attain an overall classification accuracy of 96.4% (216/224).
Table 2.
Confusion matrix of 259 testing slides for diagnosing frozen sections of thyroid nodules. The overall accuracy of all testing slides is 83.4%. Both benign and malignant categories have high precision, which is 95.3% and 96.7%, respectively, while the sensitivity of uncertain category is 100%.
| Ground truth | Prediction | ||||
|---|---|---|---|---|---|
| Benign | Uncertain | Malignant | Overall | Sensitivity | |
| Benign | 61 | 19 | 5 | 85 | 71.8% |
| Uncertain | 0 | 7 | 0 | 7 | 100% |
| Malignant | 3 | 16 | 148 | 167 | 88.6% |
| Overall | 64 | 42 | 153 | 259 | – |
| Precision | 95.3% | 16.7% | 96.7% | – | – |
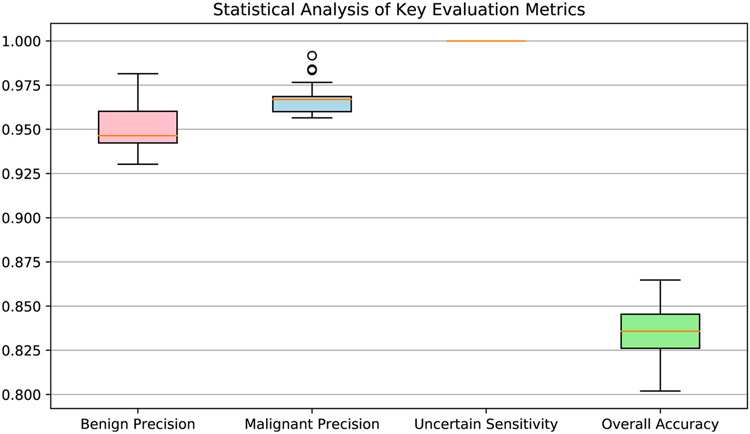
Besides the performance analysis on the all testing slides, we also conduct statistical analysis for the performance evaluation. In the statistical analysis, we randomly select 80% testing slides in each experiment. For each trial, we mainly calculate four interested metrics, including benign precision, malignant precision, uncertain sensitivity, and the overall accuracy. We conduct the trials on randomly selected slides for 100 times and draw the box plot of the results in Fig. 10. The lowest value of benign precision and malignant precision is higher than 0.925 and 0.950, respectively. As no uncertain cases are misclassified to other categories with this proposed system; thus, the sensitivity of the uncertain category is 1.0. These results satisfy the requirement of the conservative manner in thyroid frozen section diagnosis.
Fig. 10.
Statistical analysis of the four most interested metrics in thyroid frozen section diagnosis. Four interested thyroid diagnosis metrics, including benign precision, malignant precision, uncertain sensitivity, and overall accuracy, are computed via slides random sampling from the whole testing slides.
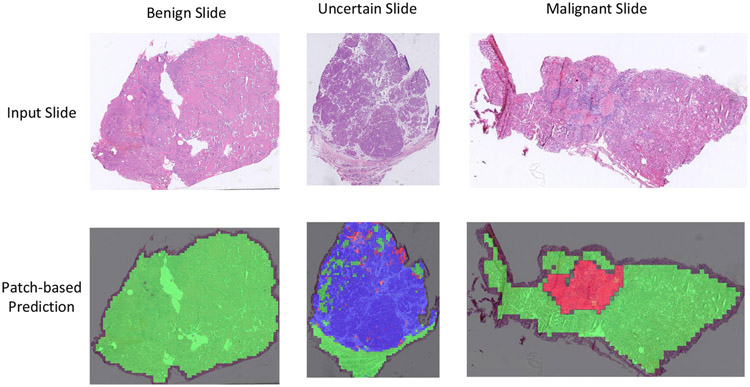
With the patch-based slide diagnosis, corresponding regions for each category are also generated along with the patch classification. Examples of suspicious region localization are shown in Fig. 9, which combines three binary maps.
Fig. 9.
Example predictions of patch-based classification of thyroid frozen sections. The first row shows benign, uncertain, and malignant input slides. The second row shows corresponding patch-based predictions. Three predicted binary maps (benign, uncertain, and malignant) are combined into a single image, in which green represents benign; blue, uncertain; and red, malignant.
The time needed for the system to diagnose slides is mainly depended on the number of patches that need to be predicted within each slide. Thus, the width and height of the WSI and the area of tissue regions within the slide are the primary factors for the computational time cost. The diagnosis of testing slides is carried out using a GeForce GTX 1080Ti Nvidia GPU. On the 259 testing slides, it takes 220 min in total, giving an average time of 51 s per slide.
5. Discussion
5.1. Benign misclassified as malignant
In the patch fusion process, we impose demanding conditions for the system to diagnose individual patches as well as slides to be malignant. These requirements emulate the tendency of pathologists to lean towards a diagnosis of uncertain when they have any doubt, to trigger further consultation or preparation of a permanent tissue section before undertaking potentially unnecessary surgery. As a result of these strict criteria, most misclassified slides are predicted to be uncertain.
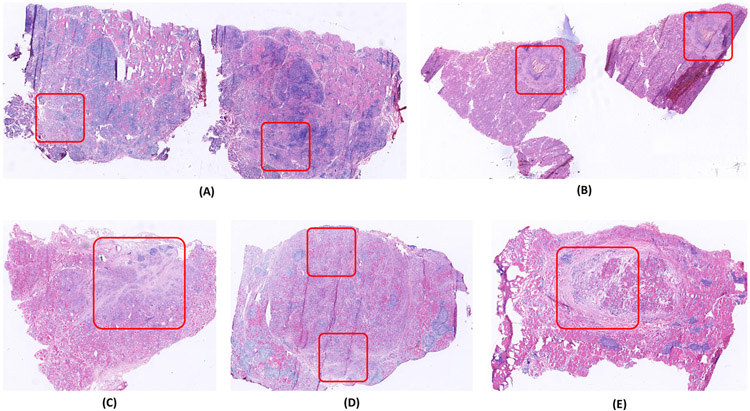
Nevertheless, there are still five benign slides in the testing set that are misclassified as malignant. The results may reflect, in part, inadequate exposure to the variety of histological variations, even though the model is trained and evaluated with more than 300 frozen sections. It also reflects that benign lesions share some features with malignant ones. The case of lymphocytic thyroiditis (Fig. 11(A)) and the case of Hashimoto thyroiditis (Fig. 11(D)) may have been misclassified as malignant because they show focally enlarged nuclei similar to nuclei in papillary thyroid carcinoma [33]. The other three misclassified cases (Fig. 11(B), (C) and (E)) are multinodular goiters that show fibrosis, hemosiderin deposition, or fibrotic encapsulation. These degenerative features also occur in malignant lesions [46]. These misclassifications also demonstrate the complexity of diagnosing thyroid frozen sections.
Fig. 11.
Five benign slides that the proposed system misclassifies as malignant. Those regions demarcated using the red bounding boxes, appear very similar to the malignant regions on malignant thyroid slides.
5.2. Patch size selection
The performance of the proposed system is mainly based on the prediction accuracy of the patch classification model, which is affected by the size of the cropped patch to a large extent. Here we choose a patch size of 2392 × 2392 pixels as a trade-off between the advantages and disadvantages of smaller and larger patches. A larger patch size means fewer patches in the slide, which can speed up diagnosis but reduce the diversity of samples from which the patch classification model can learn. Furthermore, since the input image size for InceptionV3 is 299 × 299 pixels, a larger patch size leads to a significant loss of image information. As for smaller patch size, it will prolong computations and reduce the amount of contextual information needed for diagnosis. With the chosen patch size, we can directly load slide image with the size 299 × 299 from the 3rd level of the slide, which corresponds to image patch size of 2392 × 2392 in original level 0 slide. In addition, the chosen patch size allows the model to diagnose a single thyroid slide within 1 min for most cases, satisfying the time requirements. More importantly, pathologists can obtain enough information on this patch size to make confident diagnoses. From the viewpoint of simulating pathologists’ way to make a diagnosis, this patch size should also be feasible for the CNN model.
5.3. Patch model improvement
The accuracy of the current patch classification model is lower than 0.8, which requires further improvement. The following three measures can be helpful to improve patch model performance. First, there exist noisy labels in the patches cropped through ROI-based annotations. On the one hand, we can refine the ROI annotations to be more precise. On the other hand, we can check the cropped patches and validate its labels. However, both methods are very time-consuming. Second, we can continue to collect more slide samples to improve model's robustness and generalization ability, especially uncommon frozen sections, such as Hashimoto thyroiditis, granulomatous thyroiditis, medullary carcinoma, poorly differentiated thyroid carcinoma, and thyroid lymphoma; and cases with atypical features or diagnostic challenges, such as multinodular goiter with prominent fibrosis or papillary structures (papillary hyperplasia), and thyroiditis with cytological atypia. Third, with the aim of applying the system to clinical settings, it may be possible to supplement this initial training with “feedback” training in which the model is confronted with the slides that are misclassified and thus used to refine the classification model accordingly.
5.4. Transferring to other tissues
Deep learning is a data-driven approach that discriminative features of patches can be learned when sufficiently diverse samples are collected. In this study, we transfer the parameters learned from the natural image to the thyroid frozen sections. Despite the difference between natural images and digital frozen sections, the experiments demonstrate the effectiveness of transfer learning. Because frozen sections of different tissues appear more similar compared with natural images, other frozen section tissue models can be transferred from the thyroid patch classification model instead of being transferred from the model trained from ImageNet, which may help reduce the required number of annotations and facilitate the development of automatic diagnosis system for other tissues.
For histopathology tissue diagnosis, it is essential to clearly define diagnostic categories before the collection of slides. For thyroid nodules, the possible categories are relatively well-defined (benign, uncertain, and malignant). Other tissue with more complex situations may require greater effort and guidance from pathologists and surgeons.
5.5. Assistant to pathologists
Here we demonstrate that deep learning can contribute to the diagnosis of thyroid nodules from intraoperative frozen sections, which may allow more reliable and faster decision-making about the best surgical strategy to follow. We envisage that the proposed deep learning-based diagnosis system can be a valuable assistant to pathologists. Instead of diagnosing based on the slide alone, the pathologist may refer to the automatic diagnosis and focus on suspicious areas flagged by the model. The system may identify regions that the pathologist missed, and it may reinforce the pathologist's thinking by providing complete information for the more reliable diagnosis, and even lead the pathologist to seek a second opinion. This system may be particularly useful in environments where suitably qualified pathologists are lacking. At this stage, deep learning can complement, but not replace, a certified pathologist.
6. Conclusions
In this paper, we present a patch-based system to automatically diagnose thyroid nodules from intraoperative frozen sections using deep learning techniques. With all patches’ predictions using the InceptionV3 model, we develop a rule-based protocol to fuse all predictions for the diagnosis of thyroid nodules, which allow for interpretability for the diagnosis. The proposed system achieves high precision on benign and malignant categories of thyroid nodules of 95.3% and 96.7%, respectively. In addition, a typical whole slide image can be diagnosed within 1 min. These results demonstrate the accuracy and efficiency of the proposed system. In future work, we will refine the system for thyroid nodule diagnosis mainly by improving the patch classification performance and extend to frozen sections of other organs, such as lung nodules and ovarian tumors.
Acknowledgment
We thank Yong Jiang and Shuhao Wu for the assistant on data collection and management.
Footnotes
Conflict of interests
The authors declare no conflicts of interest.
References
- [1].Novis DA, Gephardt GN, Zarbo RJ. Interinstitutional comparison of frozen section consultation in small hospitals: a college of American pathologists Q-probes study of 18532 frozen section consultation diagnoses in 233 small hospitals. Arch Pathol Lab Med 1996;120(12):1087. [PubMed] [Google Scholar]
- [2].Collins KA. The future of the forensic pathology workforce. Acad Forens Pathol 2015;5(4):526–33. 10.23907/2015.058. [DOI] [Google Scholar]
- [3].Robboy SJ, Weintraub S, Horvath AE, Jensen BW, Alexander CB, Fody EP, et al. Pathologist workforce in the United States: I. Development of a predictive model to examine factors influencing supply. Arch Pathol Lab Med 2013;137(12):1723–32. 10.5858/arpa.2013-0200-OA. [DOI] [PubMed] [Google Scholar]
- [4].Benediktsson H, Whitelaw J, Roy I. Pathology services in developing countries: a challenge. Arch Pathol Lab Med 2007;131(11):1636–9. [DOI] [PubMed] [Google Scholar]
- [5].Field A. Training for cytotechnologists and cytopathologists in the developing world. Cytopathology 2016;27(5):313–6. 10.1111/cyt.12372. [DOI] [PubMed] [Google Scholar]
- [6].Chen J, Jiao Y, Lu C, Zhou J, Zhang Z, Zhou C. A nationwide telepathology consultation and quality control program in China: implementation and result analysis. Diagnostic pathology, Vol. 9 2014:S2. 10.1186/1746-1596-9-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dong Y, Bai J, Zhang Y, Shang G, Zhao Y, Li S, et al. Automated quantitative cytology imaging analysis system in cervical cancer screening in Shanxi Province, China. Cancer Clin Oncol 2017;6(2):51–9. 10.5539/cco.v6n2p51. [DOI] [Google Scholar]
- [8].Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60–88. 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- [9].He B, Guan Y, Dai R. Classifying medical relations in clinical text via convolutional neural networks. Artif Intell Med 2019;93:43–9. 10.1016/j.artmed.2018.05.001. [DOI] [PubMed] [Google Scholar]
- [10].Bui TD, Lee J-J, Shin J. Incorporated region detection and classification using deep convolutional networks for bone age assessment. Artif Intell Med 2019;97:1–8. 10.1016/j.artmed.2019.04.005. [DOI] [PubMed] [Google Scholar]
- [11].Zhang Z, Chen P, Shi X, Yang L. Text-guided neural network training for image recognition in natural scenes and medicine. IEEE Trans Pattern Anal Mach Intell 2019. 10.1109/TPAMI.2019.2955476. [DOI] [PubMed] [Google Scholar]
- [12].Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. Imagenet large scale visual recognition challenge. Int J Comput Vis 2015;115(3):211–52. 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- [13].He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition 2016:770–8. 10.1109/CVPR.2016.90. [DOI] [Google Scholar]
- [14].Silver D, Huang A, Maddison CJ, Guez A, Sifre L, Van Den Driessche G, et al. Mastering the game of go with deep neural networks and tree search. Nature 2016;529(7587):484. 10.1038/nature16961. [DOI] [PubMed] [Google Scholar]
- [15].Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Advances in neural information processing systems 2012:1097–105. 10.1145/3065386. [DOI] [Google Scholar]
- [16].Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. Proceedings of the IEEE conference on computer vision and pattern recognition 2015:3431–40. 10.1109/CVPR.2015.7298965. [DOI] [PubMed] [Google Scholar]
- [17].Ren S, He K, Girshick R, Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. Advances in neural information processing systems 2015:91–9. 10.1109/TPAMI.2016.2577031. [DOI] [PubMed] [Google Scholar]
- [18].Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542(7639):115. 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016;316(22):2402–10. 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- [20].Wang H, Roa AC, Basavanhally AN, Gilmore HL, Shih N, Feldman M, et al. Mitosis detection in breast cancer pathology images by combining handcrafted and convolutional neural network features. J Med Imaging 2014;1(3):034003. 10.1117/1.JMI.1.3.034003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ertosun MG, Rubin DL. Automated grading of gliomas using deep learning in digital pathology images: a modular approach with ensemble of convolutional neural networks. AMIA annual symposium proceedings, Vol. 2015 2015:1899. [PMC free article] [PubMed] [Google Scholar]
- [22].Cruz-Roa A, Gilmore H, Basavanhally A, Feldman M, Ganesan S, Shih NN, et al. Accurate and reproducible invasive breast cancer detection in whole-slide images: a deep learning approach for quantifying tumor extent. Sci Rep 2017;7:46450. 10.1038/srep46450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bejnordi BE, Veta M, Van Diest PJ, Van Ginneken B, Karssemeijer N, Litjens G, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017;318(22):2199–210. 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Xu J, Luo X, Wang G, Gilmore H, Madabhushi A. A deep convolutional neural network for segmenting and classifying epithelial and stromal regions in histopathological images. Neurocomputing 2016;191:214–23. 10.1016/j.neucom.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Serag A, Hamilton P, Maxwell P, O’Reilly P. A multi-level deep learning algorithm to estimate tumor content and cellularity of prostate cancer. 2018. [Google Scholar]
- [26].Xing F, Yang L. Robust nucleus/cell detection and segmentation in digital pathology and microscopy images: a comprehensive review. IEEE Rev Biomed Eng 2016;9:234–63. 10.1109/RBME.2016.2515127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shi X, Xing F, Xu K, Xie Y, Su H, Yang L. Supervised graph hashing for histopathology image retrieval and classification. Med Image Anal 2017;42:117–28. 10.1016/j.media.2017.07.009. [DOI] [PubMed] [Google Scholar]
- [28].Xie Y, Xing F, Shi X, Kong X, Su H, Yang L. Efficient and robust cell detection: a structured regression approach. Med Image Anal 2018;44:245–54. 10.1016/j.media.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Xie Y, Chen P, Cai J, Xing F, Yang L. Efficient and robust cell segmentation in breast microscopy image using fully convolutional neural network with multi-context aggregation. Lab Invest 2018;98:113. [Google Scholar]
- [30].Xing F, Xie Y, Shi X, Chen P, Zhang Z, Yang L. Towards pixel-to-pixel deep nucleus detection in microscopy images. BMC Bioinf 2019;20(1):1–16. 10.1186/s12859-019-3037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kahmke R, Lee WT, Puscas L, Scher RL, Shealy MJ, Burch WM, et al. Utility of intraoperative frozen sections during thyroid surgery. Int J Otolaryngol 2013. 10.1155/2013/496138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Guevara N, Lassalle S, Benaim G, Sadoul J-L, Santini J, Hofman P. Role of frozen section analysis in nodular thyroid pathology. Eur Ann Otorhinolaryngol Head Neck Dis 2015;132(2):67–70. 10.1016/j.anorl.2014.02.006. [DOI] [PubMed] [Google Scholar]
- [33].Albores-Saavedra J, Wu J. The many faces and mimics of papillary thyroid carcinoma. Endocr Pathol 2006;17(1):1–18. [DOI] [PubMed] [Google Scholar]
- [34].Cho U, Mete O, Kim M-H, Bae JS, Jung CK. Molecular correlates and rate of lymph node metastasis of non-invasive follicular thyroid neoplasm with papillary-like nuclear features and invasive follicular variant papillary thyroid carcinoma: the impact of rigid criteria to distinguish non-invasive follicular thyroid neoplasm with papillary-like nuclear features. Mod Pathol 2017;30(6):810. 10.1038/modpathol.2017.9. [DOI] [PubMed] [Google Scholar]
- [35].Borrelli N, Denaro M, Ugolini C, Poma AM, Miccoli M, Vitti P, et al. Mirna expression profiling of ’noninvasive follicular thyroid neoplasms with papillary-like nuclear features’ compared with adenomas and infiltrative follicular variants of papillary thyroid carcinomas. Mod Pathol 2017;30(1):39. 10.1038/modpathol.2016.157. [DOI] [PubMed] [Google Scholar]
- [36].Hou L, Samaras D, Kurc TM, Gao Y, Davis JE, Saltz JH. Patch-based convolutional neural network for whole slide tissue image classification. Proceedings of the IEEE conference on computer vision and pattern recognition 2016:2424–33. 10.1109/CVPR.2016.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chen P, Xie Y, Hai S, Yang L. Automatic pathology diagnosis on large slide image using patch aggregation. Lab Invest; 2018. p. 586. [Google Scholar]
- [38].Wei JW, Tafe LJ, Linnik YA, Vaickus LJ, Tomita N, Hassanpour S. Pathologist-level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks. Sci Rep 2019;9(1):3358. 10.1038/s41598-019-40041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhang Z, Chen P, McGough M, Xing F, Wang C, Bui M, et al. Pathologist-level in-terpretable whole-slide cancer diagnosis with deep learning. Nat Mach Intell 2019; 1 (5): 236–45. 10.1038/s42256-019-0052-1. [DOI] [Google Scholar]
- [40].Wang X, Chen H, Gan C, Lin H, Dou Q, Tsougenis E, et al. Weakly supervised deep learning for whole slide lung cancer image analysis. IEEE Trans Cybern 2019. 10.1109/TCYB.2019.2935141. [DOI] [PubMed] [Google Scholar]
- [41].Ilse M, Tomczak JM, Welling M. Attention-based deep multiple instance learning. 2018. arXiv preprint arXiv:1802.04712. [Google Scholar]
- [42].Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. 2014. 10.1109/ACPR.2015.7486599. arXiv preprint arXiv: 1409.1556. [DOI] [Google Scholar]
- [43].Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. Proceedings of the IEEE conference on computer vision and pattern recognition 2016:2818–26. 10.1109/CVPR.2016.308. [DOI] [Google Scholar]
- [44].Chen P, Yang L. Tissueloc: whole slide digital pathology image tissue localization. J Open Source Softw 2019;4(33):1148. 10.21105/joss.01148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ruder S. An overview of gradient descent optimization algorithms. 2016. arXiv preprint arXiv:1609.04747. [Google Scholar]
- [46].Nishiyama RH. Overview of surgical pathology of the thyroid gland. World J Surg 2000;24(8):898–906. 10.1007/s002680010157. [DOI] [PubMed] [Google Scholar]