Figure 2.
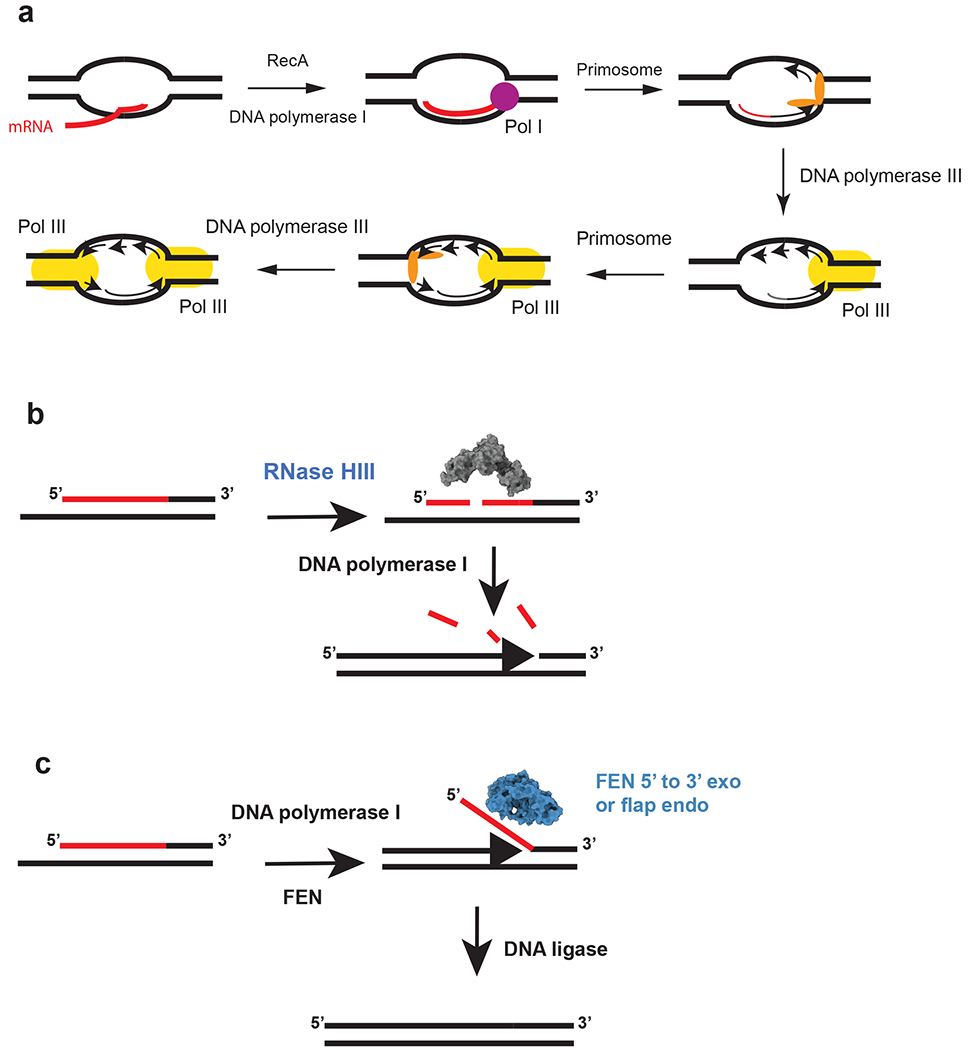
Initiation of replication from R-loops and a model for Okazaki fragment processing in RNase HIII–containing bacteria. The red lines indicate RNA, and the black lines denote DNA. (a) Model for cSDR. The nascent transcript is paired with DNA by RecA in negative supercoiled DNA behind RNAP. The transcript primes synthesis by DNA polymerase I followed by primosome assembly and loading of DNA polymerase III. The second fork is activated following primosome assembly and loading of DNA polymerase III. This figure is based on models from the following References 15 and 41 (b) RNase HIII incises the RNA portion of the Okazaki fragment. The incisions allow for DNA polymerase I to efficiently remove the RNA during DNA synthesis from the 3′-OH on the adjacent fragment (86). (c) Bacteria that have a stand-alone FEN can cleave the flap resulting from DNA polymerase I strand displacement synthesis. For bacteria that contain RNase HIII and FEN, it is expected that both function during Okazaki fragment maturation (1, 86). The model is based on the following reference (86). The space filling models for RNase HIII and FEN were generated using the B. subtilis protein sequences modeled with I-TASSER (116). Abbreviation: FEN, flap endonuclease/5′-to-3′ exonuclease.
