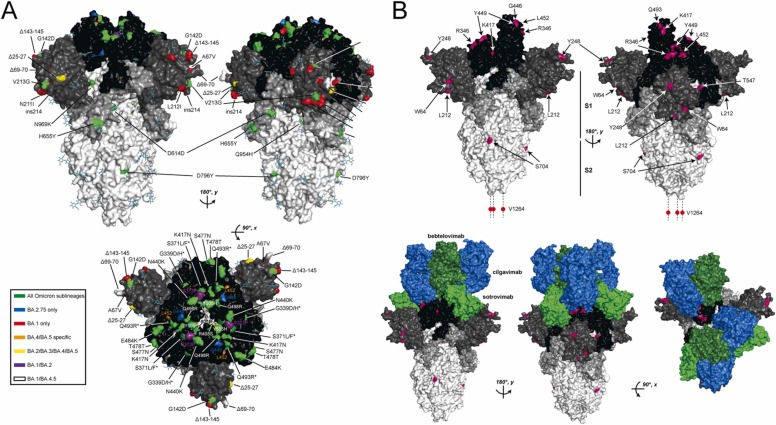
Fig. 1.
SARS-CoV-2 S protein mutations found in Omicron variant sublineages. A) Positions on the S protein where amino acid mutations are found in different Omicron sublineages are mapped to the spike trimer structure (PBD 6VXX). The S trimer is rotated 180 degrees along the y-axis to show both faces of the long axis and 90 degrees about the x-axis to display mutations on the receptor binding domains. Positions with amino acid mutations found in all Omicron sublineages are colored green. Positions that are BA.1 and BA.2.75 strain-specific are colored blue and red, respectively, and mutations found only in BA.4/BA.5, BA.2/BA.3/BA.4/BA.5, BA.1/BA.2 and BA.1/BA.2 are colored orange, yellow, purple and white, respectively. Positions where amino acid substitutions are different across different sublineages are indicated with an asterisk. B) Instances of convergent evolution in the Omicron sublineages are highlighted in magenta on the homotrimeric S protein structure (PBD 7C2L). The domain organization of S is highlighted with RBD, NTD, S1, and S2 subunits colored black, dark grey, light grey, and white, respectively. The homotrimer is rotated 180 degrees about the y-axis to show both faces. In the top panel, the sites of convergent evolution on the S homotrimer alone are indicated. In the bottom panel, the interfaces with clinically important mAbs from 3 different classes (Focosi et al., 2022c) (cilgavimab, bebtelovimab, and sotrovimab; colored blue, dark green and light green, respectively) are displayed to highlight the importance of the positions of convergent evolution in the Omicron S protein.