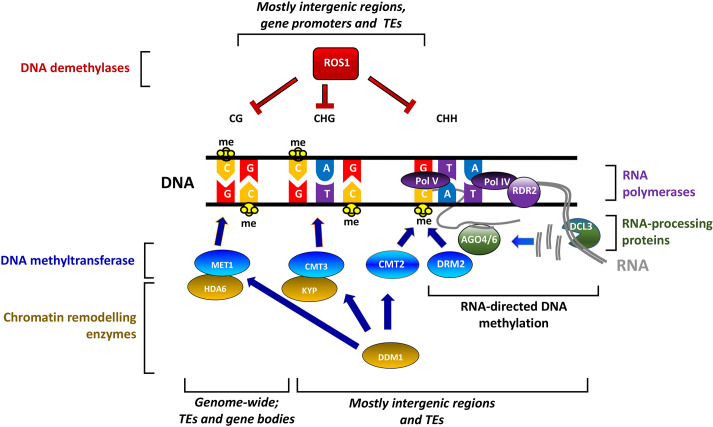
Figure 3. DNA methylation homeostasis in plants.
DNA methylation predominantly occurs at the 5-carbon of cytosine and typically signals for tightly packed chromatin (heterochromatin) to prevent transcription by RNA polymerase II. DNA methylation in plants occurs at three different sequence contexts: CG, CHG and CHH where H stands for any nucleotide aside from guanine [4]. While DNA methylation is most prevalent at transposable elements (TEs), it occurs throughout the plant genome, including gene bodies [47]. In Arabidopsis, the induction and establishment of cytosine methylation is mediated RNA-directed DNA methylation (RdDM), which involves RNA polymerases IV and V, small interfering RNAs (siRNAs), AGO (Argonaute) proteins and the DNA methyltransferase DRM2, which methylates DNA at CHH context [48,49]. Not shown are variations to the RdDM pathway that initiate CHH methylation (often referred to as non-canonical RdDM, which involves RNA pol II, AGO1 and RDR6 [50]). Once established, methylation at CG sequence context is maintained by MET1 (Methyltransferase 1), while the methyltransferases CMT2 and CMT3 (Chromomethylase 2/3) maintain methylation at CHH and CHG contexts, respectively [51]. These DNA methyl transferases often interact directly or indirectly with chromatin remodelling enzymes to ensure tightly packed heterochromatin. Demethylation is executed by four demethylases, DME (Demeter), DML2 (Demeter-Like 2), DML3 (Demeter-Like 3) and ROS1 (Repressor Of Silencing 1), of which the latter is expressed in vegetative tissues [52]. The antagonistic activities between the DNA demethylases and the methylation pathways determines the level of DNA methylation at transposable elements [53].