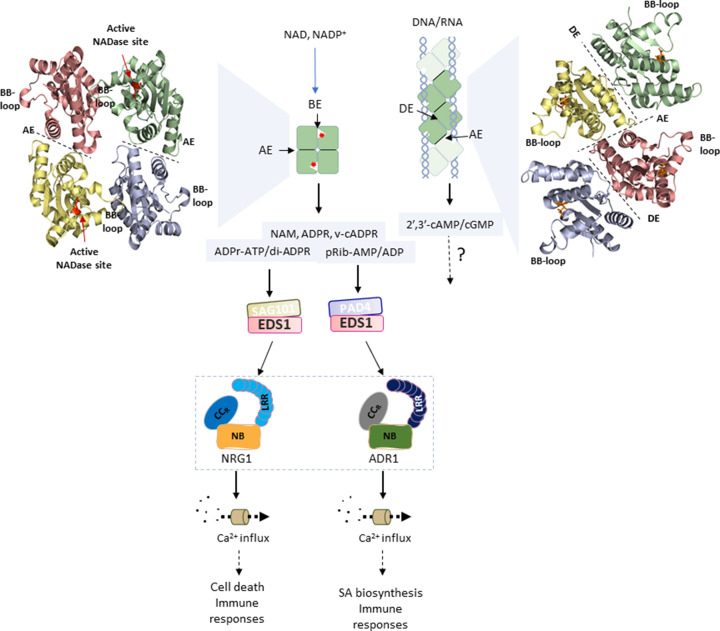
Figure 3. Working model for TIR activation and signalling pathways.
Upon TIR-NLR activation, resistosome formation causes the TIR domains (indicated by the green squares in the schematic model) to oligomerize through AE and BE interfaces to activate NADase activity, which cleaves NAD or NADP+ to generate various small molecules and ADP-ribosylation products (left). Alternatively, TIR domains may also self-associate through AE and DE interfaces to form helical filaments, which can bind and hydrolyze DNA/RNA and generate 2′,3′-cAMP/cGMP (right). Extreme left: Roq1TIR tetramer cryo-EM structure (PDB: 7JLX). Extreme right: L6TIR (PDB: 3OZI) tetramer structural model assembled through AE and DE interfaces. The TIR monomer subunit structures are shown in green, yellow, lavender, and salmon with the catalytic glutamate residues shown in red in the resistosome tetramer (left) and in orange in the AE–DE filament (right). The NADase activity products ADPr-ATP/di-ADPR bind to EDS1/SAG101 complexes, while pRibAMP/ADP binds to EDS1/PAD4 complexes. This results in activation of their helper CCR-NLRs, NRG1, and ADR1, respectively. NRG1s promote cell death, whereas ADR1s induce SA biosynthesis, achieved by triggering Ca2+ influx or through some other unknown mechanism(s). Solid arrows indicate confirmed links, dotted arrows indicate tentative pathways.