Extended Data Figure 7. Comparison of naïve RorcCreZbtb46f/f mice relative to littermates.
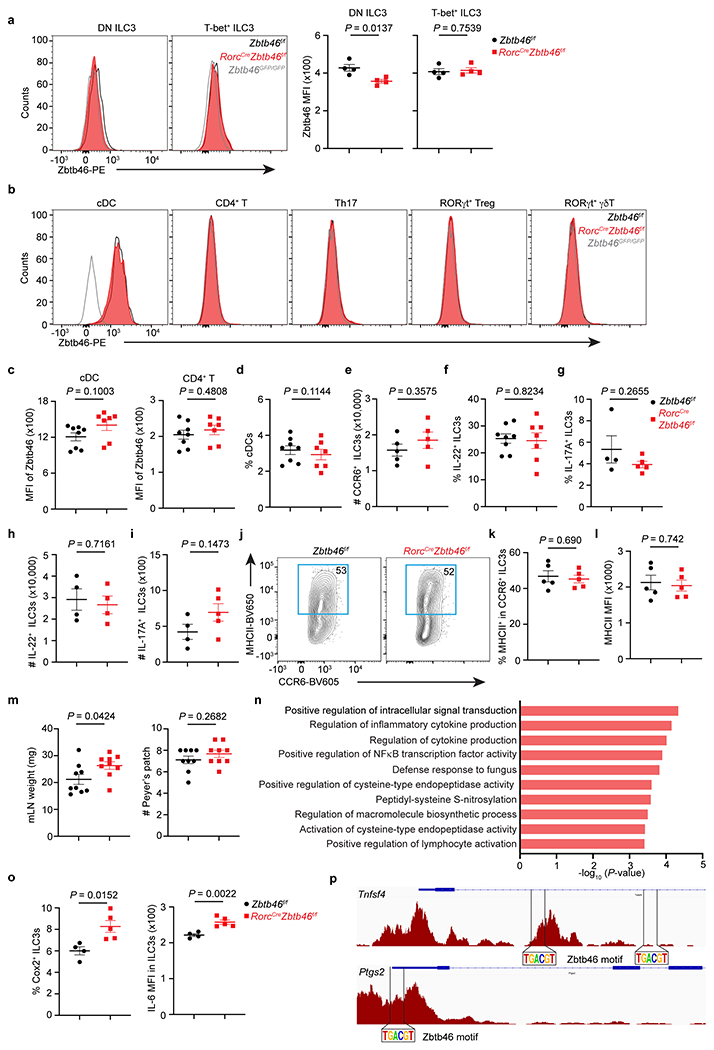
a. Representative flow cytometry histograms and quantification of Zbtb46 MFI in double-negative (DN) or T-bet+ ILC3s from the large intestine of indicated mice (n = 4 mice per group for both subsets). b. Representative flow cytometry histograms of Zbtb46 in cDCs and different T cell subsets in the large intestine of indicated mice. c. Quantification of geometric MFI of Zbtb46 in cDCs and CD4+ T cells (n = 8 or 7 mice per group for both cell types). d. The frequency of cDCs in the large intestine of indicated mice (n = 8 or 7 mice per group). e. Quantification of CCR6+ ILC3 cell numbers in the large intestine of indicated mice (n = 5 mice per group). f. Quantification of the frequencies of IL-22+ ILC3s in indicated mice (n = 8 or 7 mice per group). g. Quantification of the frequencies of IL-17A+ ILC3s in indicated mice (n = 4 or 5 mice per group). h. Quantification of the cell numbers of IL-22+ ILC3s in indicated mice (n = 4 mice per group). i. Quantification of the cell numbers of IL-17A+ ILC3s (n = 4 or 5 mice per group) in indicated mice. j,k. Representative flow plots and quantification of the frequencies of MHCII+ CCR6+ ILC3s in the large intestine of indicated mice (n = 5 mice per group). l. Quantification of MHCII MFI in CCR6+ ILC3s in indicated mice (n = 5 mice per group). m. Quantification of mLN weight and the numbers of Peyer’s patches in indicated mice (n = 9 mice per group). n. Go pathway analysis of the upregulated genes in the RorcCreZbtb46f/f mice compared to Zbtb46f/f mice. o. Quantification of the frequencies of COX-2+ (n = 4 or 5 mice per group) or IL-6 MFI (n = 4 or 5 mice per group) in ILC3s in the large intestine of indicated mice. p. Zbtb46 motif analysis of the Tnfsf4 and Ptgs2 loci in ATAC-seq data from CCR6+ ILC3s. Data are representative of three independent experiments in a, b, e, g-l, o. Data are pooled from two independent experiments in c, d, f, m. Data are shown as the means ± S.E.M. Statistics are calculated by two-tailed unpaired Student’s t-test.
