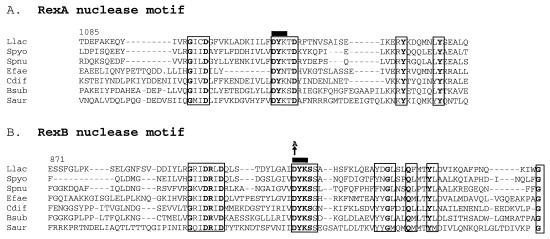
FIG. 1.
Alignment of nuclease motifs present in each subunit of the two-subunit exo/hel enzymes. (A) RexA subunit homologue alignments are presented for the region surrounding the nuclease motif (corresponding to positions 1085 to 1164 of the 1,173-amino acid L. lactis RexA subunit). (B) RexB subunit homologies in the region surrounding the nuclease motif (corresponding to positions 871 to 949 of the 1,099-amino acid L. lactis RexB subunit). Highly conserved motifs are enclosed in a rectangle, and amino acids that are totally conserved are in bold. The black bar over amino acids DYK corresponds to the region deleted in lactococcal RexA or RexB mutants; the arrow over position 910 in RexB indicates a point substitution from aspartic acid to alanine generated in RexB (see the text). Llac, L. lactis; Spyo, Streptococcus pyogenes; Spnu, Streptococcus pneumoniae; Efae, Enterococcus faecalis; Cdif, Clostridium difficile; Bsub, B. subtilis; Saur, Staphylococcus aureus.