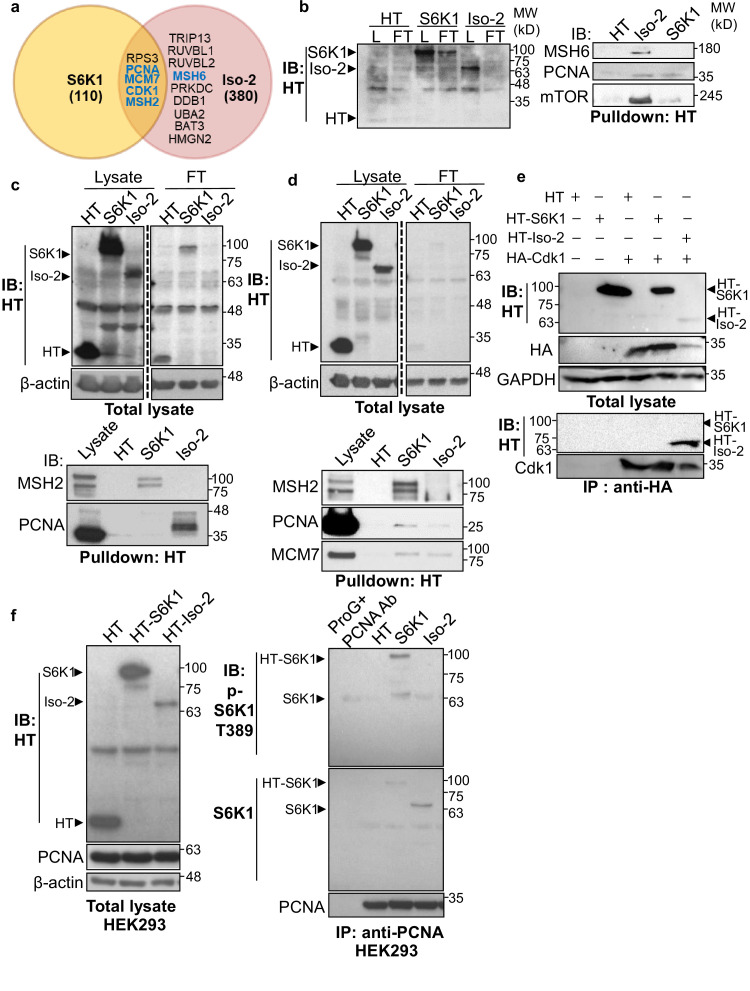
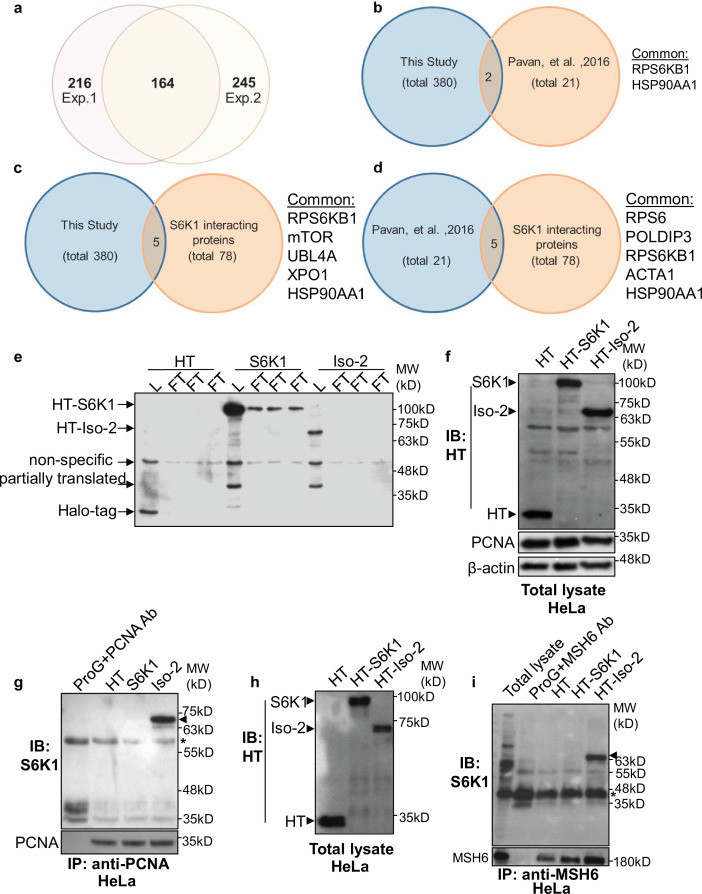
Figure 2. S6K1 and Iso-2 bind to DNA damage/repair proteins.
(a) Venn diagram representing DNA damage response/repair proteins that bound specifically to HT-S6K1 isoforms in transfected HEK293 cells. Proteins marked in blue were validated by reciprocal immunoprecipitations. (b,c) Western blot of total lysate (L) and flow through (FT) of Halo-tag pulldown from HEK293 cells transfected with either Halo-tag expressing control vector (HT), HT-S6K1 (S6K1) or HT-Iso-2 (Iso-2) (b, left panel and c, upper panel). Western blot of Halo-tag pulldown from the same cells (b, right panel c, lower panel). (d) Western blot of total lysate (L) and flow through (FT) of Halo-tag pulldown from HeLa cells transfected with either Halo-tag expressing control vector (HT), HT-S6K1 (S6K1) or HT-Iso-2 (Iso-2) (upper panel). Western blot of Halo-tag pulldown from the same cells transfected with either Halo-tag expressing control vector (HT), S6K1 or Iso-2 (lower panel). (e) Western blot of total lysate from HEK293 cells co-transfected with either Halo-tag expressing control vector (HT), HT-S6K1 (S6K1) or HT-Iso-2 (Iso-2) and HA-Cdk1 (upper panel). Western blot of immunoprecipitation of lysates of these cells with anti-HA antibody (lower panel). (f). Western blot analysis of HEK293 cells transfected as described in b (left panel). Western blot of immunoprecipitation of lysates of these cells with anti-PCNA antibody (right panel).