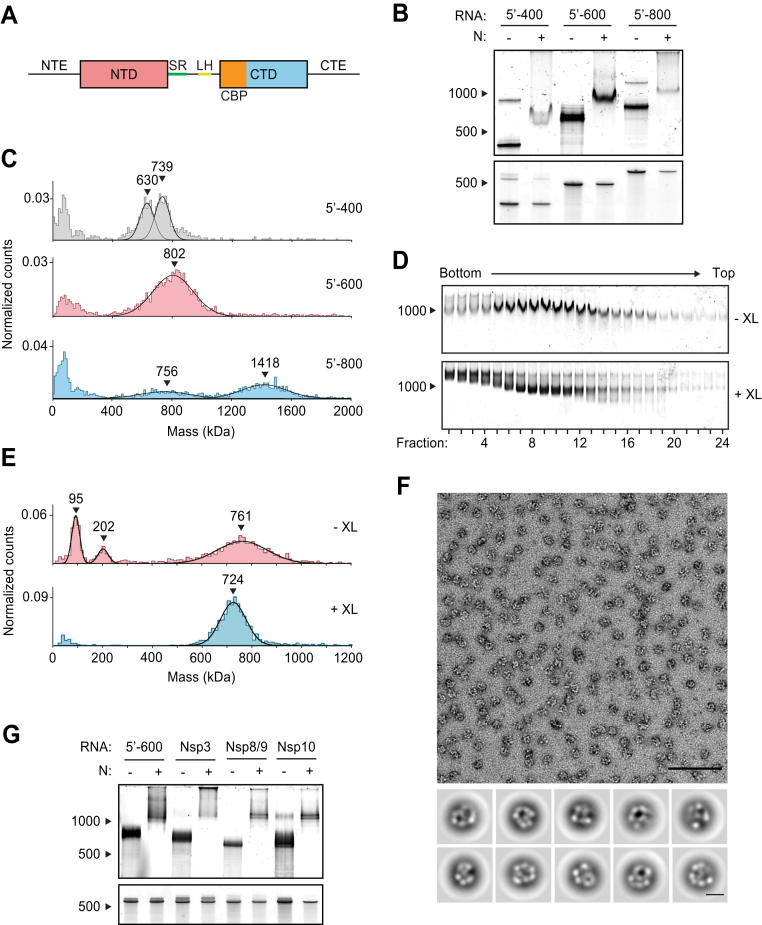
Figure 1.
Viral RNA promotes formation of the SARS-CoV-2 ribonucleosome.A, schematic of N protein domain architecture, including the N-terminal extension (NTE), N-terminal domain (NTD), serine/arginine region (SR), leucine helix (LH), C-terminal basic patch (CBP), C-terminal domain (CTD), and C-terminal extension (CTE). B, native (top) and denaturing (bottom) PAGE analysis of 15 μM N protein mixed with 256 ng/μl of the indicated RNA, stained with SYBR Gold to detect RNA species. RNA length standards shown on left (nt). RNA concentration in these and other experiments was 256 ng/μl, regardless of RNA length, to ensure that all mixtures contain the same nucleotide concentration. C, mass photometry analysis of vRNP complexes formed in the presence of 15 μM N and 256 ng/μl RNA. Data were fit to Gaussian distributions, with mean molecular mass indicated above each peak. Representative of two independent experiments (Table S1). D, native gel analysis of glycerol gradient separated vRNP complexes. Top, no crosslinker added (−XL); bottom: 0.1% glutaraldehyde added (+XL) to 40% glycerol buffer (GraFix). RNA length standard shown on left (nt). E, fractions 7 and 8 (from D) were combined and analyzed by mass photometry, as in C. Top, no crosslinker (−XL); bottom, GraFix-purified vRNP (+XL). Representative of two independent experiments (Table S1). F, negative stain electron microscopy and two-dimensional classification of GraFix-purified vRNPs (combined fractions 7 and 8 from D). Scale bars are 100 nm (top) and 10 nm (bottom). G, native (top) and denaturing gel analysis (bottom) of 15 μM N protein mixed with 256 ng/μl of the indicated 600 nt RNA molecules. RNA length standards shown on left (nt). See Table S2 for sequences. N, nucleocapsid; vRNP, viral ribonucleoprotein.