Figure 2.
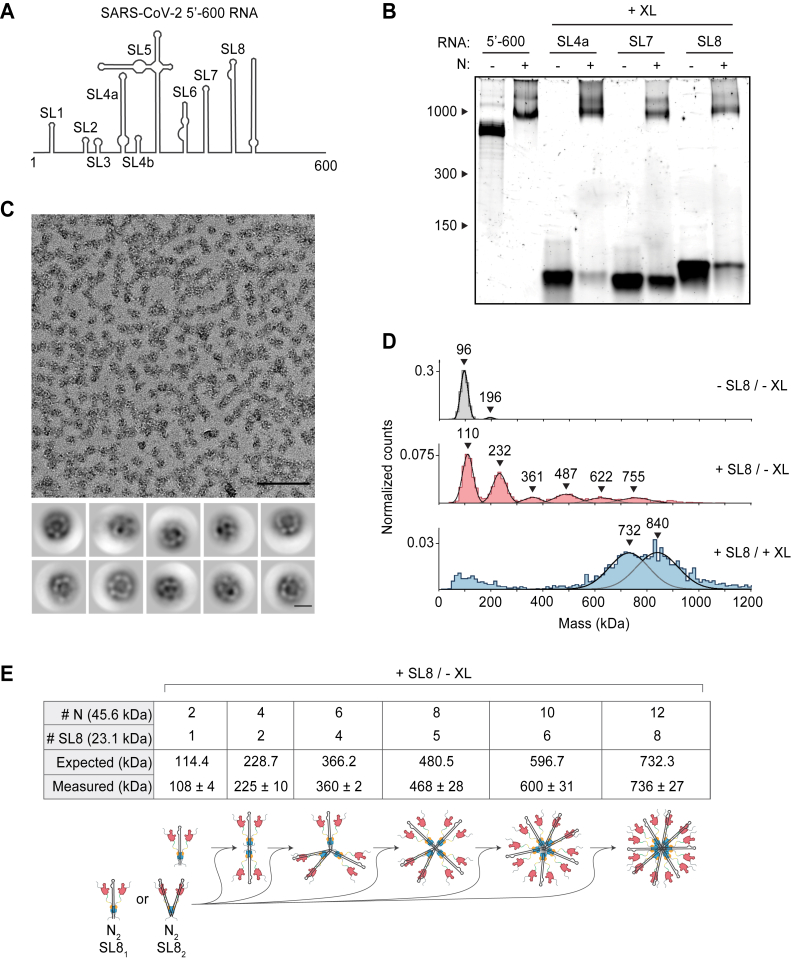
Stem-loop RNA, in complex with N protein, drives ribonucleosome formation.A, schematic of RNA secondary structure in the 5′-600 RNA (46). B, native gel analysis of 15 μM N protein mixed with 256 ng/μl of the indicated RNAs. Samples containing stem-loop RNA were crosslinked to stabilize the resulting complex, while the 5′-600 RNA sample was left un-crosslinked. RNA length standards shown on left (nt). Corresponding denaturing gel analysis shown in Fig. S1A. C, fractions 7 and 8 of GraFix-purified SL8 assembled vRNPs were combined and analyzed by negative stain electron microscopy and two-dimensional classification. Scale bars are 100 nm (top) and 10 nm (bottom). D, mass photometry analysis of indicated N protein-RNA mixtures. Top, N protein alone; middle, N protein mixed with SL8, un-crosslinked; bottom, crosslinked complexes of N protein bound to SL8 (data reproduced from Fig. S1B for ease of comparison). Representative of two independent experiments (Table S1). E, predictions of N protein and RNA stoichiometry, based on measured masses of N protein in complex with SL8 RNA without crosslinker (D, middle panel). Measured masses are means ± standard deviation in two independent experiments (Table S1). Below the table is a schematic of a proposed assembly mechanism in which N protein dimers, bound to one or two stem-loop RNAs, iteratively assemble to the full vRNP. N, nucleocapsid; vRNP, viral ribonucleoprotein.