Figure 5.
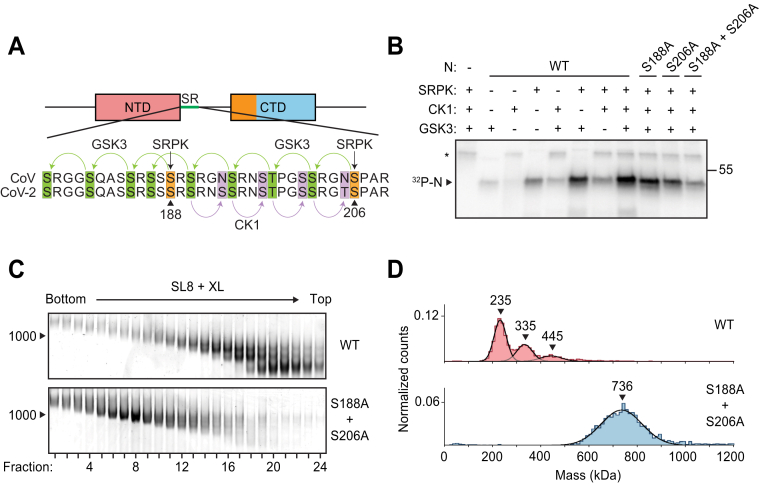
Phosphorylation of N protein inhibits ribonucleosome formation.A, sequence of N protein SR regions from SARS-CoV (aa 177–210) and SARS-CoV-2 (aa 176–209). The proposed mechanism of sequential phosphorylation (30) is initiated by SRPK at S188 and S206 (orange), which leads to upstream phosphorylation of eight sites by GSK3 (green), allowing for final phosphorylation of four additional sites by CK1 (purple). In the phosphomimetic 10D mutant used in Figure 4, the SRPK and GSK3 sites are changed to aspartic acid. B, wildtype (WT) and mutant N protein constructs were incubated with the indicated kinases in the presence of radiolabeled ATP and analyzed by SDS-PAGE and autoradiography. Phosphorylated N is indicated. Asterisk denotes autophosphorylation of CK1. Molecular mass marker shown on right (kDa). C, N protein (WT or S188A + S206A) was phosphorylated by SRPK, GSK3, and CK1 and then mixed with SL8 RNA. The resulting ribonucleoprotein complexes were separated by glycerol gradient centrifugation in the presence of crosslinker (GraFix) and analyzed by native gel electrophoresis. RNA length standard shown on left (nt). D, peak fractions from the GraFix analyses in C were analyzed by mass photometry. Top, fractions 19 + 20 of wildtype N; bottom, fractions 7 + 8 of S188A + S206A mutant N. Representative of two independent experiments (Table S1). CK1, casein kinase 1; GSK3, glycogen-synthase kinase 3; N, nucleocapsid; SR, serine/arginine region; SRPK, serine-arginine protein kinase.