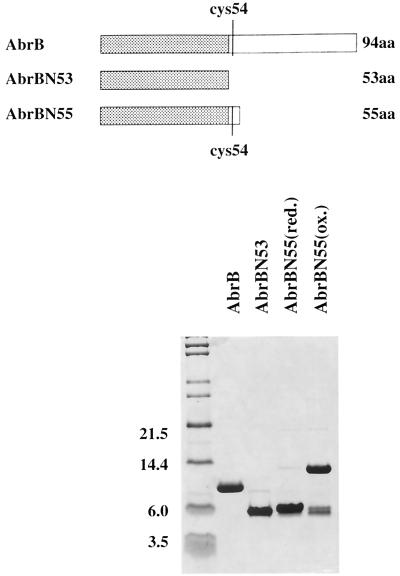
FIG. 2.
Purified proteins used in DNA-binding assays. At the top is a cartoon depicting the nature of the monomeric subunits of the proteins. The purity of the proteins used and the comparative difference between the reduced and oxidized (disulfide-linked) preparations of AbrBN55 are shown at the bottom by a sodium dodecyl sulfate-polyacrylamide gel electrophoresis fractionation. Note that the truncated proteins used in this study, as well as wild-type AbrB (16), have the sequence MKST… at their amino terminus (N terminus). However, in the original report announcing the sequence of the abrB gene (7), the reading frame was presumed to start at a Met codon located 6 bp upstream (and in frame) from the codon that specifies the Met residue at the N terminus of the pure proteins. Most of the previous literature and database sequence depositories have followed the original convention indicating that the N terminus of AbrB is MFMKST… While we cannot rule out the possibility that in vivo translation starts at the upstream Met codon, with subsequent processing of the protein to produce the observed N terminus, we believe the true in vivo start to be at the downstream Met codon. In any event, since our pure proteins had MKST… at their N terminus, we were compelled to number the residues of the protein accordingly. Unfortunately, if reference is made to the previous literature, a discrepancy of two residues will be observed in the numbering of the amino acids, e.g., the lone Cys residue in the monomer is Cys54 in this paper but was Cys56 previously. Molecular masses (in kilodaltons) are shown.