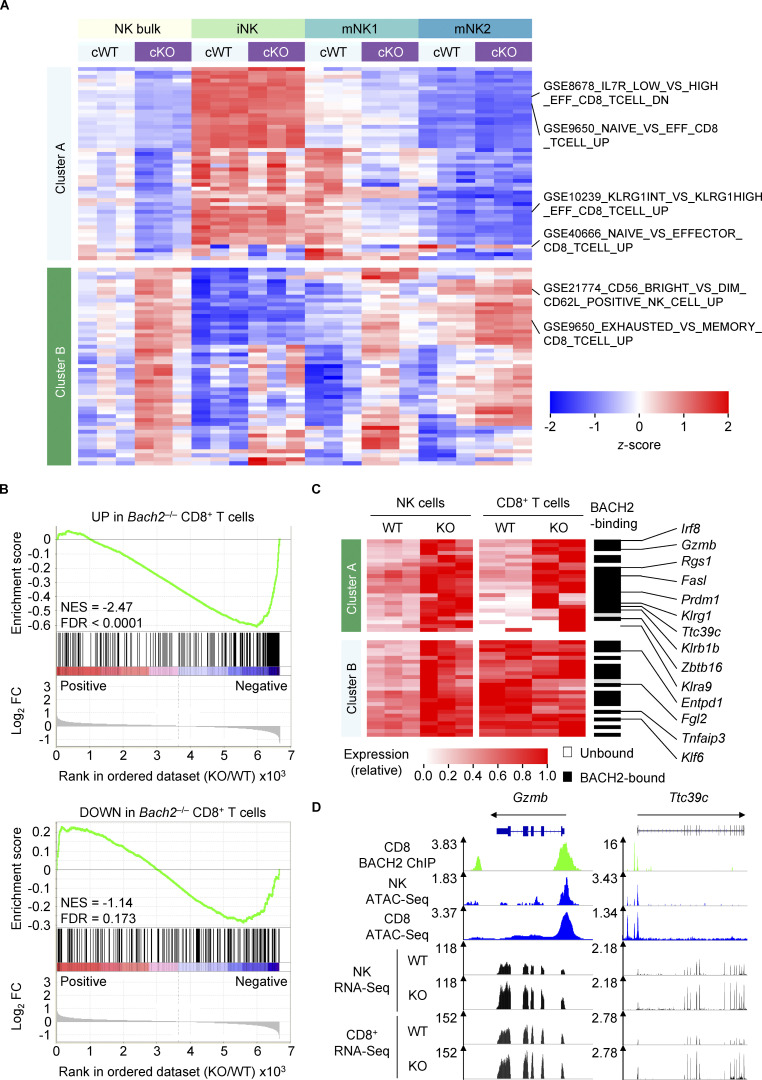
Figure 4.
BACH2 regulates shared transcriptional programs in NK and CD8+ T cells. (A) ssGSEA of cWT and cKO NK cells and subsets with immunologic gene sets (ImmuneSigDB; Pval < 0.05, |FC| > 1.5). (B) GSEA of differentially upregulated (top) and differentially downregulated (bottom) genes in Bach2−/− CD8+ T cells within the global transcriptional differences between cKO and cWT NK cells. NES, normalized enrichment score; FDR, false discovery rate. (C) Heatmap showing expression of genes which are significantly upregulated in cKO NK cells (Padj ≤ 0.2) and expressed by both NK and CD8+ T cells. Genes with peak-called BACH2-binding sites between 15 kb upstream of the transcription start site and 3 kb downstream of the transcription end site of that gene within CD8+ T cells are indicated. cWT/cKO NK and WT/KO CD8+ T cell gene expression data (reads per million; RPM) independently normalized to row maxima. Genes are hierarchically clustered along the y axis. (D) Representative alignments of ChIP-Seq, ATAC-Seq, and RNA-Seq data at selected loci in indicated cell types, showing increased mRNA expression of genes. RNA-Seq from three independent biological replicates per genotype is shown.