Fig. 6.
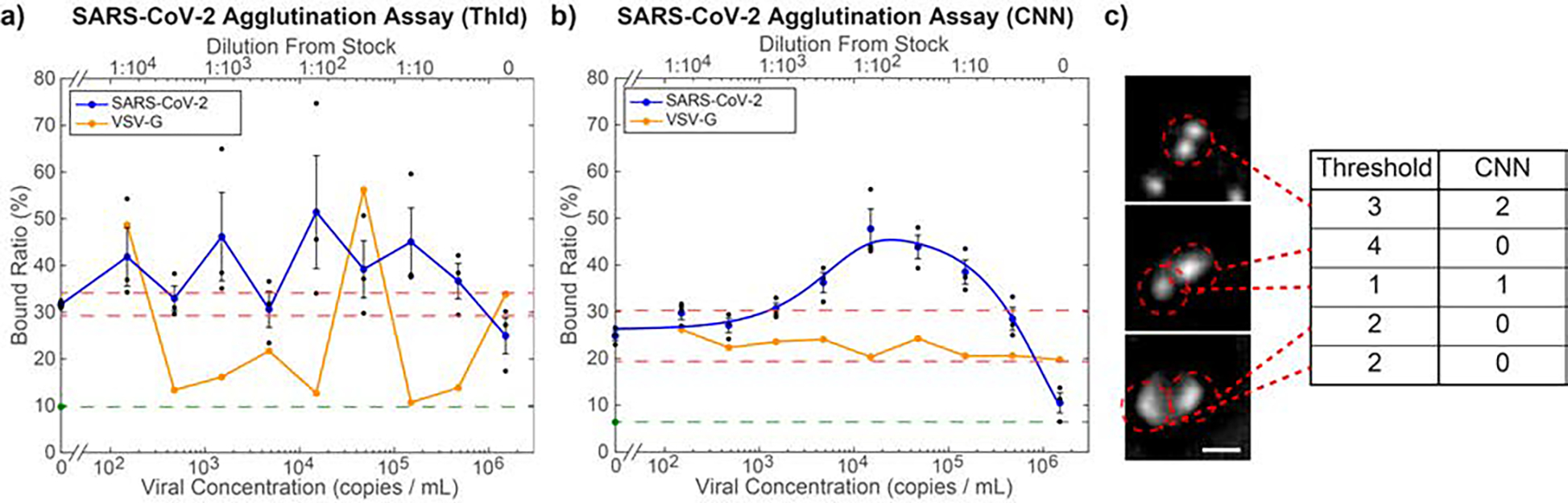
Quantification of microbead agglutination in the presence of SARS-CoV-2 pseudovirus. a) Optimized threshold-based quantification of the agglutination assay, which fails to find a clear trend in BR as a function of concentration. Over all non-control virus concentrations, the BR shows an average standard deviation of 10.66%, indicating inconsistency in the threshold-based quantification. Orange points indicate measurement results for VSV-G, which is used as a negative control. As the viral concentration of the VSV-G was not independently measured, the dilutions of the sample relative to the stock solution (top axis) were matched to the same dilutions of the SARS-CoV-2 pseudovirus, whose stock concentration was independently measured, yielding the bottom axis values. b) CNN-based agglutination quantification. Average standard of deviation for the BR of all non-control virus concentrations is 3.89%. The blue curve is a best fit to Eq. 5 (R2 = 0.974). The LOD based on this curve is 1,270 viral copies·mL−1. Orange points are negative control measurements using VSV-G samples diluted as specified in panel (a). The lack of a significant response shows that this assay is specific to SARS-CoV-2. For both graphs, the red dashed lines are the upper and lower LOD cutoffs, while the green point and dashed line indicate the BR in pure PBS. Black points indicate triplicate samples for each concentration and error bars are standard error of the mean. c) Example images classified by thresholding vs CNN. The CNN correctly identified each one of these features, while thresholding did not. Scale bar = 5 μm.
