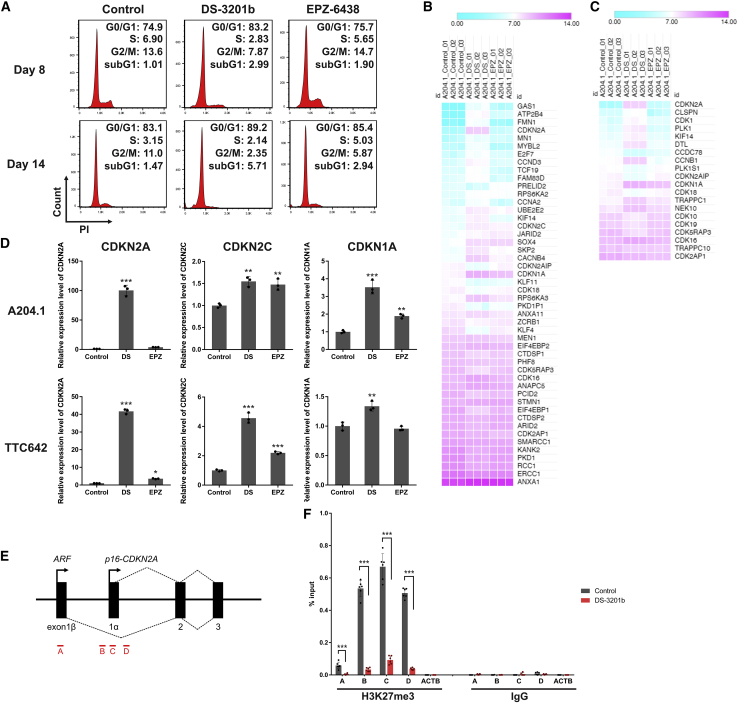
Figure 5.
The reactivation of CDKN2A, one of the EZH1/2 targets, and induction of cell-cycle arrest by EZH1/2 dual inhibition
(A) Representative histograms of cell-cycle analysis. A204.1 cells were treated with DS-3201b (100 nM) or EPZ-6438 (100 nM) for the indicated duration, followed by the cell-cycle analysis with PI-staining. The y axes represent cell numbers, and x axes show DNA content (PI intensity). The numbers indicate the percentages of cells in each cell-cycle phase. (B and C) Heatmaps of differentially expressed genes belonging to the “G1/S transition of mitotic cell cycle” (GO: 0000082; B) and “G2/M transition of mitotic cell cycle” (GO: 0000086; C) terms. Mean values of signal intensity acquired from RNA-seq data are represented by colors (n = 3 each, false discovery rate adjusted p < 0.05, fold-change > 2). (D) Expression of CDKN2A, CDKN2C, and CDKN1A mRNAs in DS-3201b- or EPZ-6438-treated cells. Cells were treated with DS-3201b (DS; 100 nM) or EPZ-6438 (EPZ; 100 nM) for 7 days, followed by qRT-PCR. n = 3, means ± SD. The expression level of the control is indicated as “1.” ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus control. (E) Schematic of the CDKN2A locus and the locations of primer pairs for ChIP-qPCR analysis. (F) H3K27me3 enrichment at the CDKN2A locus. A204.1 cells were treated with DS-3201b (100 nM) for 7 days, followed by ChIP-qPCR analysis. Normal rabbit IgG was used as a negative control for the immunoprecipitation. The primer pair against ACTB was used as a negative control for the qPCR. n = 6, means ± SD. ∗∗∗p < 0.001, as indicated by the bracket.