Figure 1. Characteristics of P. aeruginosa G2L4 RT.
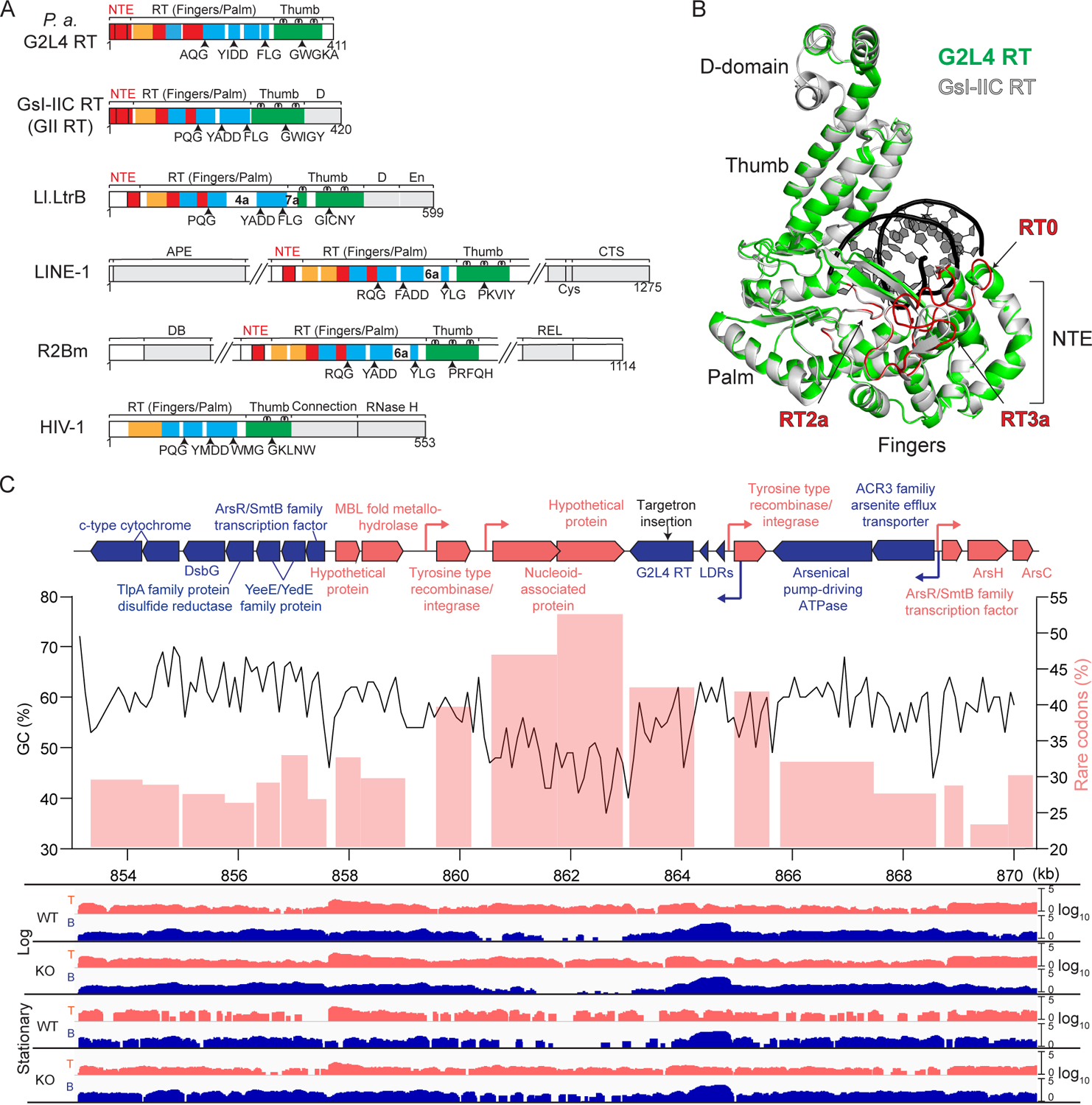
(A) Schematics comparing P. aeruginosa G2L4 RT to RTs encoded by group II introns G. stearothermophilus GsI-IIC (GII) and Lactococcus lactis Ll.LtrB (LtrA protein), human LINE-1 and Bombyx mori R2Bm non-LTR retrotransposons, and retrovirus HIV-1. Protein regions: RT1–7, conserved sequences blocks found in all RTs; NTE/RT0, RT2a and RT3a insertions relative to retroviral RTs (red); fingers (orange); palm (blue); thumb (green); other domains (gray).
(B) Three-dimensional model of G2L4 RT (green) constructed by I-TASSER (Yang and Zhang, 2015) superimposed on the crystal structure of GII RT (gray; PDB: 6AR1). Primer-template (black).
(C) Genomic region encompassing the G2L4 RT ORF in P. aeruginosa AZPAE12409. Top, map of a 17.2-kb region containing the G2L4 RT gene. Genes with protein-coding sequences on the top and bottom strand and their predicted promoters (bent arrows) are shown in red and blue, respectively. Targetron insertion site (black arrow). Middle, GC content calculated over a 500-bp sliding window (black line) and rare codon usage (pink bars) in regions around the G2L4 RT ORF. Bottom, TGIRT-seq coverage plots of cellular RNAs in the WT and G2L4 RT KO strains in log and stationary phases
