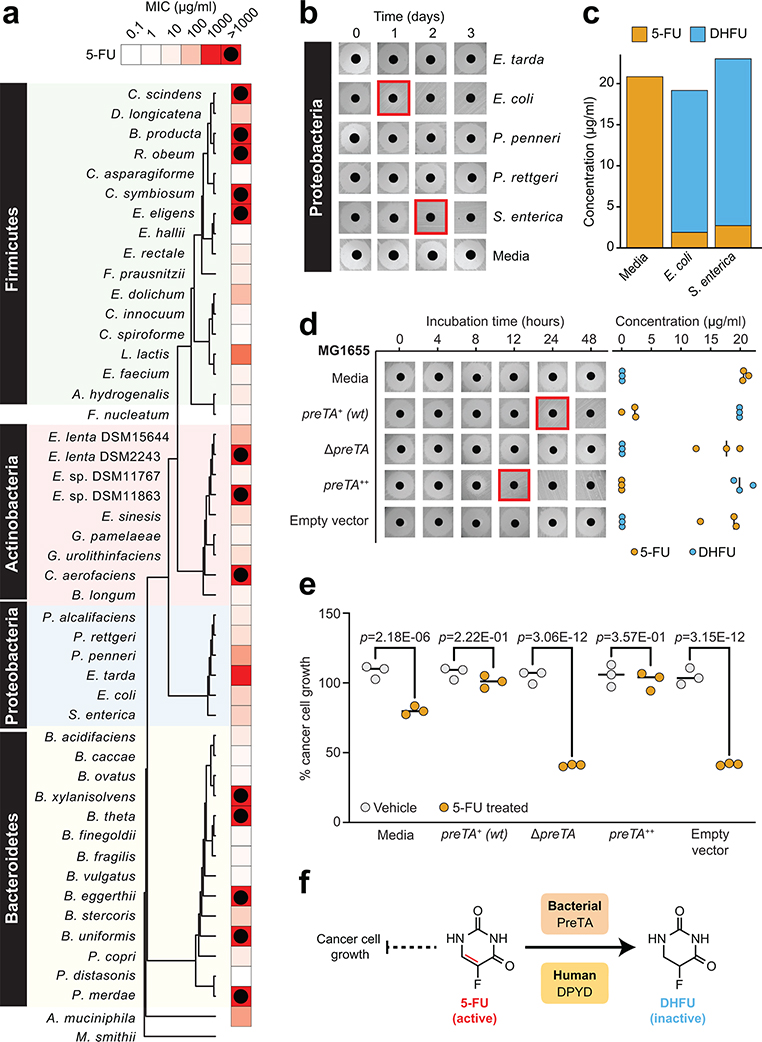
Fig. 1|. The anti-cancer drug 5-fluorouracil (5-FU) inhibits bacterial growth and is inactivated by the preTA operon.
(a) Heatmap representing the 5-FU minimal inhibitory concentration (MIC, <10% growth relative to vehicle controls; 47 human gut bacterial strains; 2 replicates/strain/concentration; see Supplementary Table 1). The phylogenetic tree was built using PhyML96, based on a MUSCLE97 alignment of full-length 16S rRNA genes. (b) We screened 23 strains with MIC≥62.5 μg/ml for drug inactivation using a disk diffusion assay (BHI+ media; 20 μg/ml 5-FU); only Proteobacteria are shown. Red squares indicate decreased zones of inhibition. (c) LC-QTOF/MS detection of the predicted 5-FU metabolite DHFU after 48 hours of anaerobic incubation [E. coli MG1655, S. enterica LT2 (DSM17058), 20 μg/ml 5-FU]. (d) Wild-type (preTA+), deletion (ΔpreTA), complemented (preTA++), and empty vector E. coli MG1655 strains were assayed for residual 5-FU using disk diffusion (0–48 hours incubation) and LC-QTOF/MS (48 hours). Complementation and empty vector were on the ΔpreTA background. Black lines indicate the median value (n=3 biological replicates). (e) The E. coli MG1655 strains shown in panel d were incubated for 72 hours with 5-FU (20 μg/ml) or vehicle (DMSO, 0.05%), conditioned media was added to the CRC cell line HCT-116, and cell proliferation was quantified using the MTT assay. Lines represent medians (n=3 biological replicates/strain/condition). p-values, one-way ANOVA with Holm-Sidak’s multiple comparisons test. (f) We propose that bacterial PreTA contributes to the elimination of 5-FU similar to hepatic expression of dihydropyrimidine dehydrogenase (DPYD).