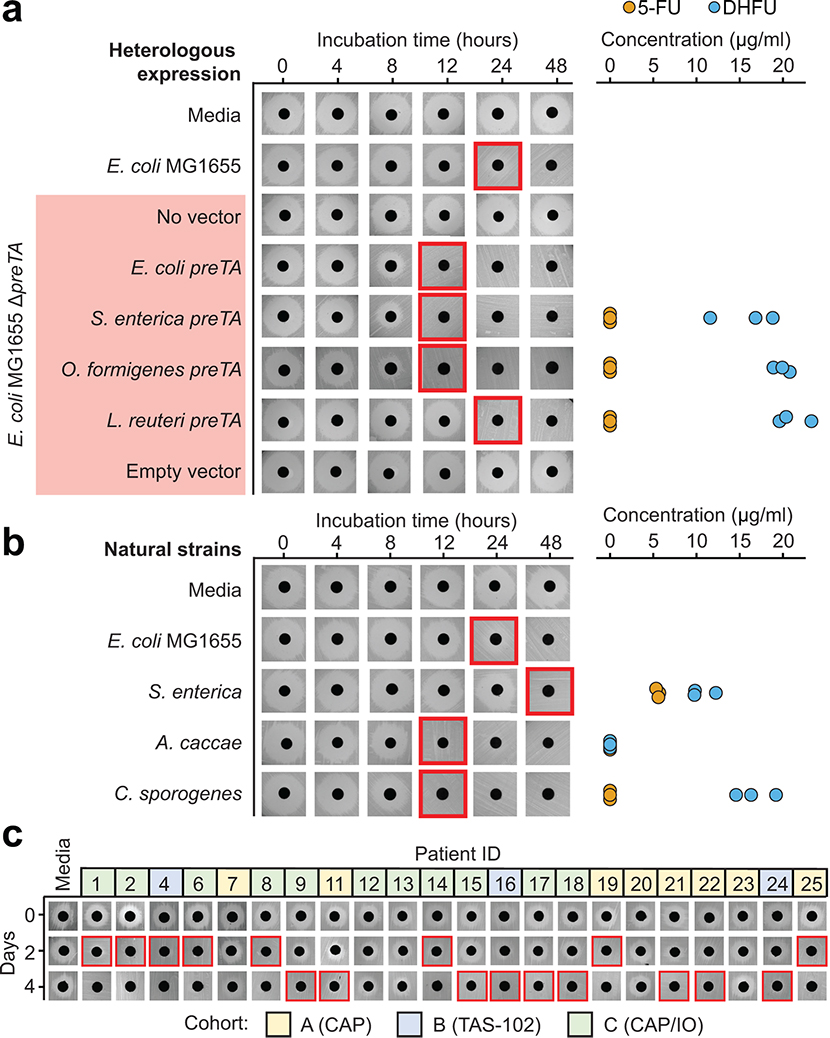
Extended Data Fig. 8. Bacterial preTA operons from diverse strains, preTA encoding natural strains, and pooled isolates from patient samples are capable of 5-FU inactivation.
(a) preTA operons from Salmonella enterica LT2 (DSM17058), Oxalobacter formigenes ATCC35274, and Lactobacillus reuteri DSM20016 were amplified and integrated into the chromosome of E. coli MG1655 ΔpreTA. Strains were incubated with 20 μg/ml 5-FU and assayed for residual drug using disk diffusion (0–48 hours incubation) and LC-QTOF/MS (48 hours). E. coli MG1655 preTA complementation and empty vector controls are also included on the ΔpreTA background. (b) We also tested strains predicted to encode the preTA operon: Salmonella enterica LT2, Anaerostipes caccae DSM14662, and Clostridium sporogenes DSM795. Residual 5-FU was assayed as in panel a. Of note, we did not detect either compound from A. caccae, suggesting that this strain may further metabolize DHFU. (c) Bacterial isolates from 22 CRC patient stool samples in the GO Study (Supplementary Table 12) were isolated on McConkey agar, pooled, and incubated in BHI+ with 5-FU for 4 days. Residual 5-FU levels were assessed using a disk diffusion assay. Colors indicate the treatment cohort: (A) CAP (yellow); (B) TAS-102 (blue); and (C) CAP plus immunotherapy (green). Red boxes in panels a-c indicate the first timepoint without a clear zone of inhibition.