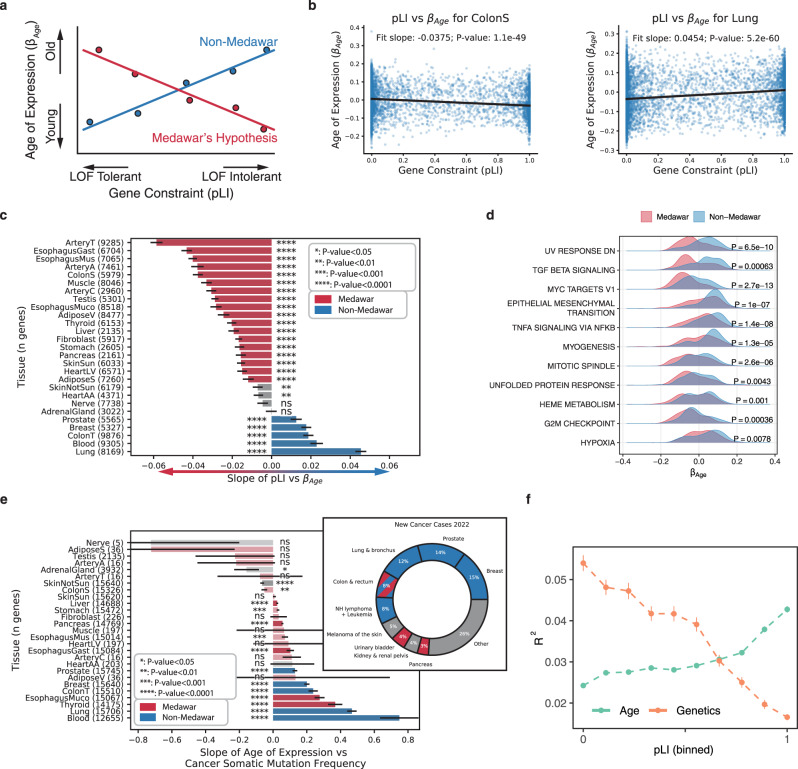
Fig. 5. Tissue-specific evolutionary signatures of aging.
a The expected relationship across genes between the per-gene age-associated slope of gene expression (βage) and a gene's level of constraint (measured by probability loss of function intolerance, pLI). Medawar's hypothesis predicts a negative relationship (shown in red) between the time of expression and the level of constraint. The opposite trend (non-Medawar) is shown in blue. b βage across genes plotted as a function of pLI for a tissue exhibiting a Medawarian signature, and a non-Medawarian signature. c The slope of the relationship from b between βage and constraint across all tissues. Error bars show estimated standard error of regression slope. Unadjusted p-values in b and c are calculated using a linear model two-sided t-test (exact p-values in Supplementary Table 2). d Hallmark pathways in which the βage was significantly different between Medawarian and non-Medawarian tissues (two-sided t-test). e Bar chart shows per-tissue relationship between βage and frequency of somatic mutations in tumor samples for a particular gene and cancer type (left). Colored as in 5C with alpha indicating unadjusted significance using two-sided t-test (exact p-values in Supplementary Table 3). Y-axis labels show the number of independent genes and error bars indicate the estimated standard error of regression slope. Doughnut chart shows estimated proportion of new cases of cancer in US in 2022 by cancer type from Table 1 of25 (right). Crosshatch indicates that while colon transverse was identified as a non-Medawar tissue, colon sigmoid was not. f Gene expression variance explained by genetics or age as a function of binned (10 bins) gene constraint averaged across all tissues. Points represent the mean and error bars the standard error of the mean.