Figure 3.
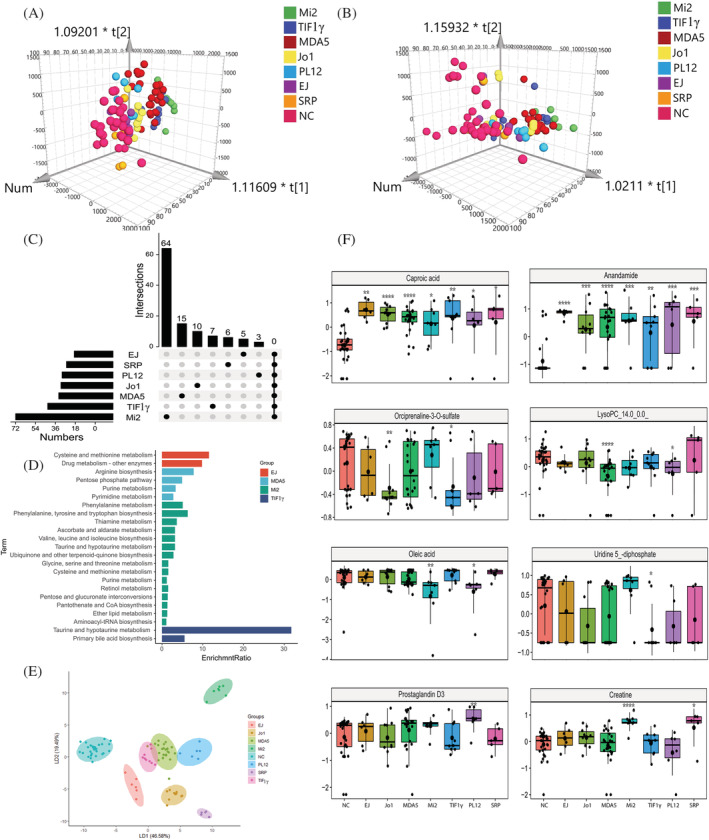
Plasma metabolomic profiles in the MSA‐defined IIM subtypes. (A and B) The Orthogonal partial least‐squares discriminate analysis (OPLS‐DA) score plots of plasma metabolomics data compared anti‐EJ+, anti‐Jo1+, anti‐MDA5+, anti‐Mi2+, anti‐TIF1γ+, anti‐PL12+, and anti‐SRP + IIM patients to normal control (NC) samples in positive (A) and negative (B) ion mode, respectively. (C) The UpSet plot analysis based on the selected important metabolites in plasma (VIP > 1, FC > 1.2 or <0.83). (D) KEGG pathway analysis of exclusively important metabolites in each IIM subtype. (E) The linear discriminant analysis (LDA) plot based on the Top 12 metabolites in random forest machine model. (F) The most specific metabolite identified by the random forest machine model to classify plasma samples from NC, anti‐EJ+, anti‐Jo1+, anti‐MDA5+, anti‐Mi2+, anti‐TIF1γ+, anti‐PL12+, and anti‐SRP + IIM patients, respectively (P value compared with NC, *, <0.05; **, <0.01; ***, <0.001, ****, <0.0001).
