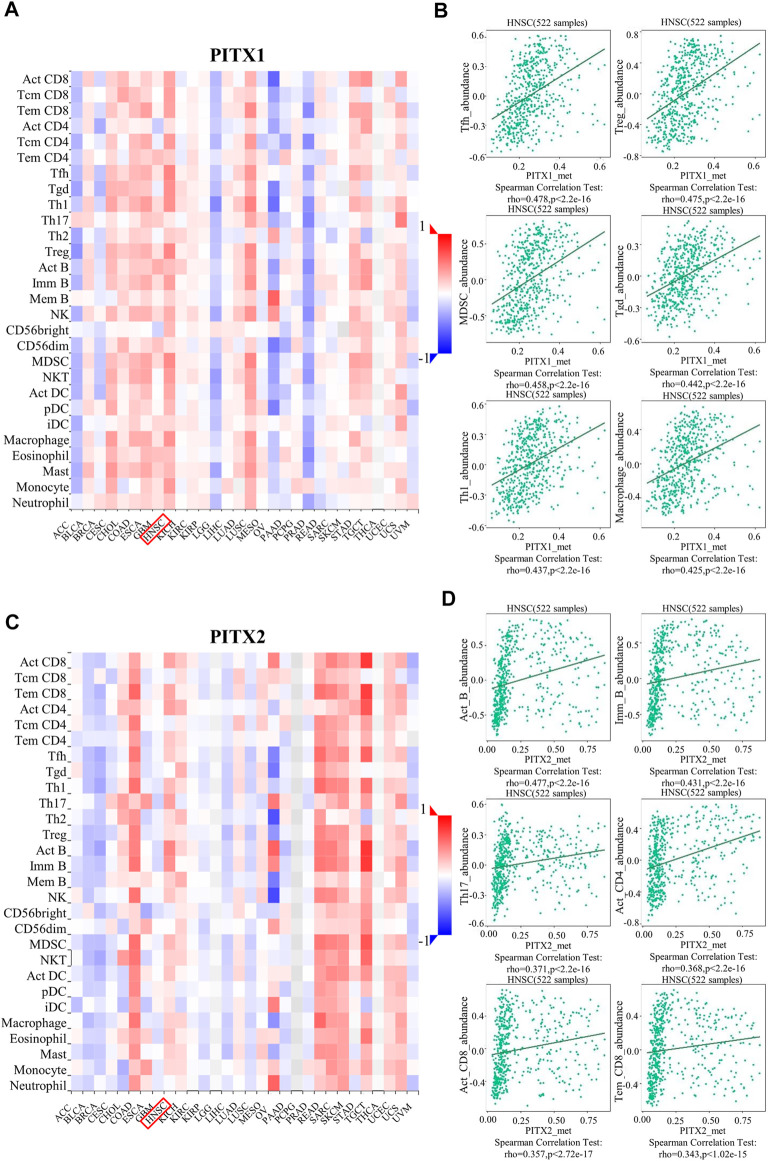
FIGURE 9.
TISIDB analysis of correlations between PITX1 and PITX2 methylation and immune cells infiltration across human cancers. (A) The relationship between PITX1 methylation and 28 types of TILs across various human cancers. (B) The top six TILs displaying the strongest Spearman’s correlation with PITX1 methylation in HNSC. (C) The relationship between PITX2 methylation and 28 types of TILs across various human cancers. (D) The top six TILs displaying the strongest Spearman’s correlation with PITX2 methylation in HNSC. Act_CD8, activated CD8+ T cells; Tcm_CD8, central memory CD8+ T cells; Tem_CD8, effector memory CD8+ T cells; Act_CD4, activated CD4+ T cells; Tcm_CD4, central memory CD4+ T cells; Tem_CD4, effector memory CD4+ T cells; Tfh, T follicular helper cells; Tgd, gamma delta T cells; Th1, type 1 T helper cells; Th17, type 17 T helper cells; Th2, type 2 T helper cells; Treg, regulatory T cells; Act B, activated B cells; Imm_B, immature B cells; Mem_B, memory B cells; NK, natural killer cells; CD56bright, CD56bright NK cells; CD56dim, CD56dim NK cells; MDSC, myeloid derived suppressor cells; NKT, natural killer T cells; Act DC, activated dendritic cells; pDC, plasmacytoid DCs; iDC, immature DCs; Mast, mast cells.