Fig. 4 |. Single Nucleus RNA Sequencing of Kidney Cortex.
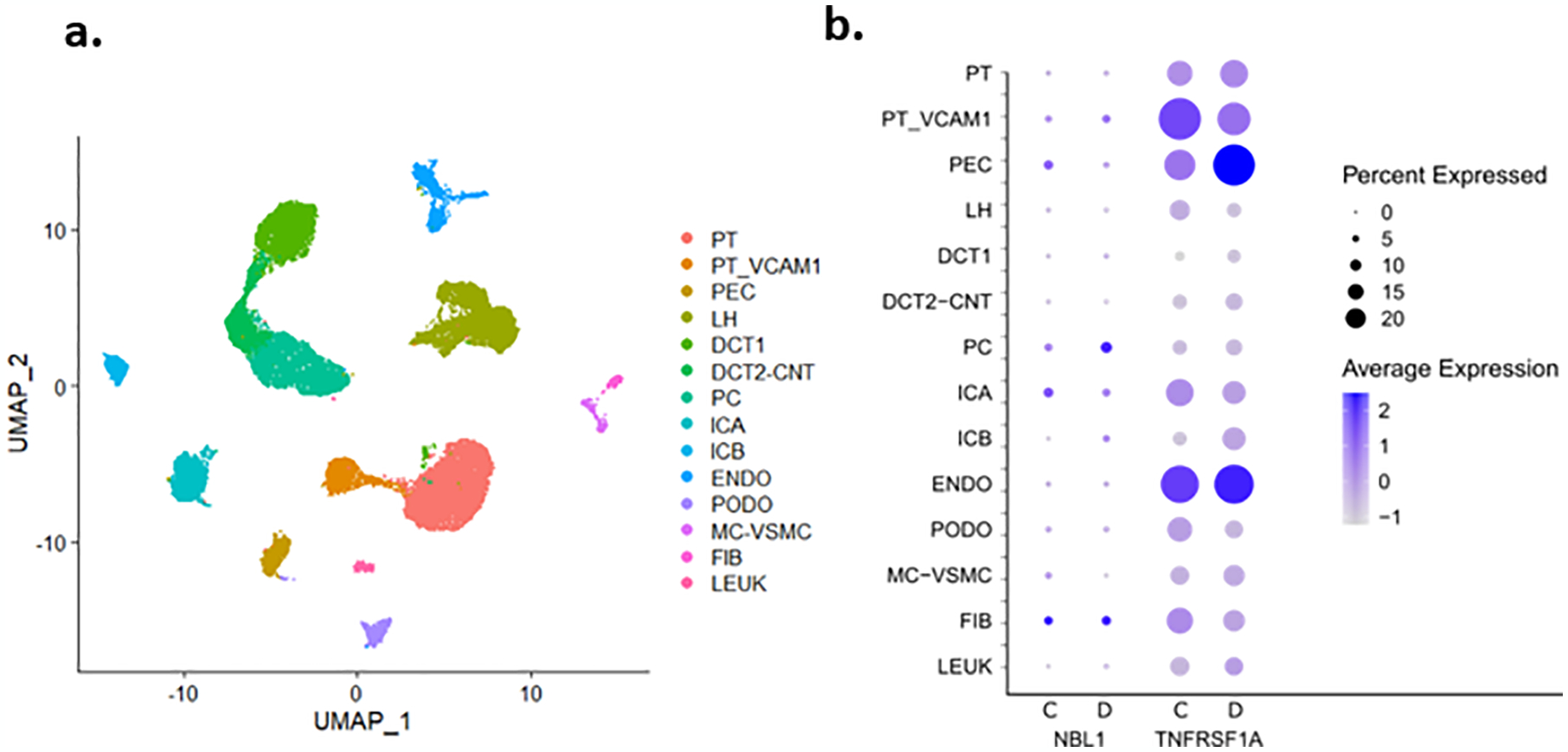
Healthy control (n=3) and patients with early diabetic kidney disease (n=3) underwent nuclear dissociation and single nucleus RNA sequencing (snRNA-seq) with 10X Genomics 5’ chemistry. Libraries were processed with cellranger and Seurat. (a) UMAP of all kidney cell types identified in the aggregated dataset. PT_VCAM1=proximal tubule cells that express VCAM1, PT=proximal tubule, PEC=parietal epithelial cells, TAL=thick ascending limb, DCT1=early distal convoluted tubule, DCT2-CNT=late distal convoluted tubule and connecting tubule, PC=principal cells, ICA=type A intercalated cells, ICB=type B intercalated cells, PODO=podocytes, ENDO=endothelial cells, MC-VSMC mesangial and vascular smooth muscle cells, FIB=fibroblasts, LEUK=leukocytes (b) NBL1 is not significantly expressed in kidney cell types in either control (C) or diabetes (D) specimens. TNFRSF1A (TNFR1) is significantly expressed in multiple kidney cell types. Scale represents normalized log-fold-change and was prepared with the Seurat DotPlot function. See Supplementary Table S3 and methods in Appendix.
