Figure 6.
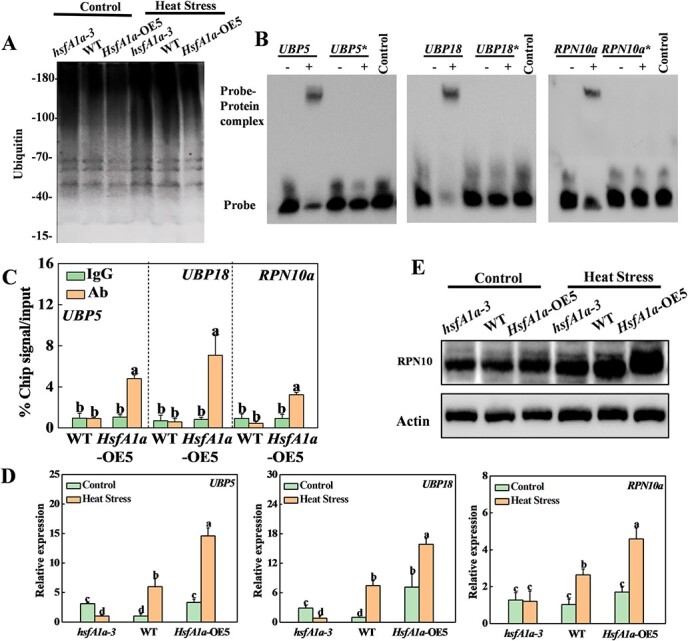
HsfA1a transcriptionally regulates ubiquitin- and proteasome-related genes in tomato anthers under heat stress. (A) Accumulation of ubiquitination for total proteins. Ubiquitinated proteins were subjected to SDS–PAGE and analyzed with an anti-ubiquitin monoclonal antibody in anthers of hsfA1a, WT, and HsfA1a-overexpressing (OE) plants. (B) EMSA analysis showing binding of HsfA1a to HSE sequences of the promoters of UBP5, UBP18, and RPN10a. Recombinant HsfA1a was used in DNA assays of binding to UBP5, UBP18, and RPN10a, and their HSE motif mutated sequences as the probes. Asterisks (*) represent mutated probes. All probes are listed in Supplementary Data Table S2. His is the negative control. (C) ChIP–qPCR was used to validate HsfA1a binding to the UBP5, UBP18, and RPN10a promoters in 35S-HsfA1a-HA-OE (HsfA1a-OE) plants. Anther samples of WT and HsfA1a-OE plants at flowering stage after heat stress were obtained, and input chromatin was isolated from them. Anti-HA antibody immunoprecipitation was used to epitope-tag the HsfA1a–chromatin complex. Mouse IgG treatment was used as a control reaction. qRT–PCR was used to quantify input and ChIP DNA samples with specific primers for the promoters of the UBP5, UBP18, and RPN10a genes. The percentages of input DNA are displayed as ChIP results. (D) Relative expression of UBP5, UBP18, and RPN10a genes in anthers of hsfA1a, WT, and HsfA1a-OE plants under the control condition or heat stress. (E) Accumulation of RPN10 proteins in anthers of hsfA1a, WT, and HsfA1a-OE plants under the control condition or heat stress. Actin was used as a loading control. Means with the same letter in Tukey’s test indicate a non-significant difference at P < .05. Similar results were obtained in three independent experiments. hsfA1a-3, one line of hsfA1a mutants; HsfA1a-OE5, one line of HsfA1a-OE plants.
