Figure 1.
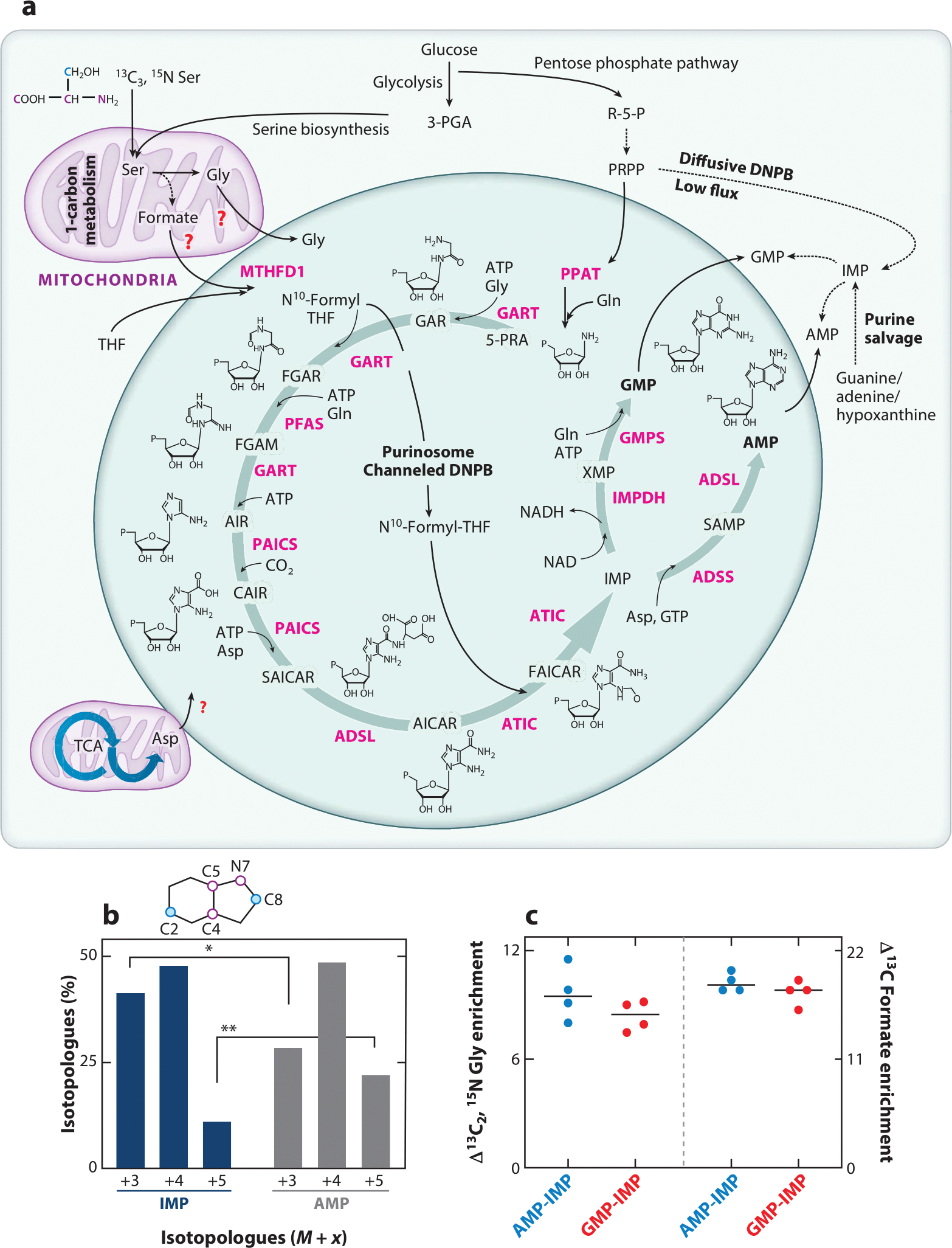
Enzymatic composition and function of the purinosome. (a) Channeled DNPB is carried out by the purinosome metabolon, which is composed of at least ten enzymes (pink): PPAT, GART, PFAS, PAICS, ADSL, ATIC, IMPDH, ADSS, GMPS, and MTHFD1. Structures of DNPB intermediates are shown; “P” denotes a phosphate group. Glycolysis, the serine biosynthesis pathway, one-carbon metabolism, the pentose phosphate pathway, and the TCA cycle generate the building block substrates utilized in DNPB. Involvement of mitochondrial Gly, formate, and Asp transporters for the direct uptake of these substrates for DNPB by purinosomes has been proposed though not confirmed yet (red question marks). The stable isotope–labeled positions of Ser are shown for the backbone atoms (pink) and side chain carbon (blue); the positions of these labeled atoms upon incorporation into the purine ring are shown as pink and blue circles in the purine ring diagram (shown in panel b). Alternatively, low-flux DNPB is carried out by the diffusive pool of DNPB enzymes outside purinosomes and by purine salvage enzymes that produce IMP, GMP, and AMP. (b) Upon 13C3, 15N Ser–mediated labeling of de novo synthesized purines, a different isotopologue distribution was discovered in the newly synthesized IMP (blue bars) and AMP (dark gray bars), revealing the two parallel purine-generating pathways (33). The purine ring positions replaced with isotope-labeled atoms are shown as pink and blue circles in the purine ring schematic. The tightly channeled, high-flux DNPB pathway that primarily generates AMP and GMP is carried out by mitochondria-associated purinosomes. Alternatively, the diffusive, low-flux DNPB primarily generates the free IMP pool. (c) The end nucleotides AMP and GMP showed higher incorporation of the mitochondrially derived substrates Gly and formate (33). AMP and GMP showed ~10% higher 13C2, 15N Gly enrichment and ~20% higher 13C formate enrichment compared with diffusively synthesized IMP. The plot shows the difference in isotope incorporation between AMP and IMP (blue circles) and GMP and IMP (red circles). Abbreviations: 3-PGA, 3-phosphoglyceric acid; 5-PRA, 5- phosphoribosylamine; ADSL, adenylosuccinate lyase; ADSS, adenylosuccinate synthetase; AICAR, 5-aminoimidazole-4-carboxamide ribonucleotide; AIR, 5-aminoimidazole ribonucleotide; ATIC, 5-aminoimidazole-4-carboxamide nucleotide formyltransferase/IMP cyclohydroxylase; CAIR, carboxyaminoimidazole ribonucleotide; DNPB, de novo purine biosynthesis; FAICAR, 5-formamidoimidazole-4-carboxamide ribonucleotide; FGAM, formylglycinamidine; FGAR, formylglycinamide ribonucleotide; GAR, glycinamide ribonucleotide; GART, phosphoribosylglycinamide formyltransferase; GMPS, GMP synthetase; IMP, inosine monophosphate; IMPDH, IMP dehydrogenase; MTHFD1, cytosolic methylenetetrahydrofolate dehydrogenase; NAD, nicotinamide adenine dinucleotide oxidized; NADH, nicotinamide adenine dinucleotide reduced; PAICS, phosphoribosyl aminoimidazole succinocarboxamide synthase; PFAS, phosphoribosyl formylglycinimidine transferase; PPAT, PRPP amidotransferase; PRPP, phosphoribosyl pyrophosphate; R-5-P, ribose-5-phosphate; SAICAR, phosphoribosyl aminoimidazole succinocarboxamide; SAMP, succinyladenosine monophosphate; TCA, tricarboxylic acid; THF, tetrahydrofolate; XMP, xanthosine monophosphate.
