Figure 3.
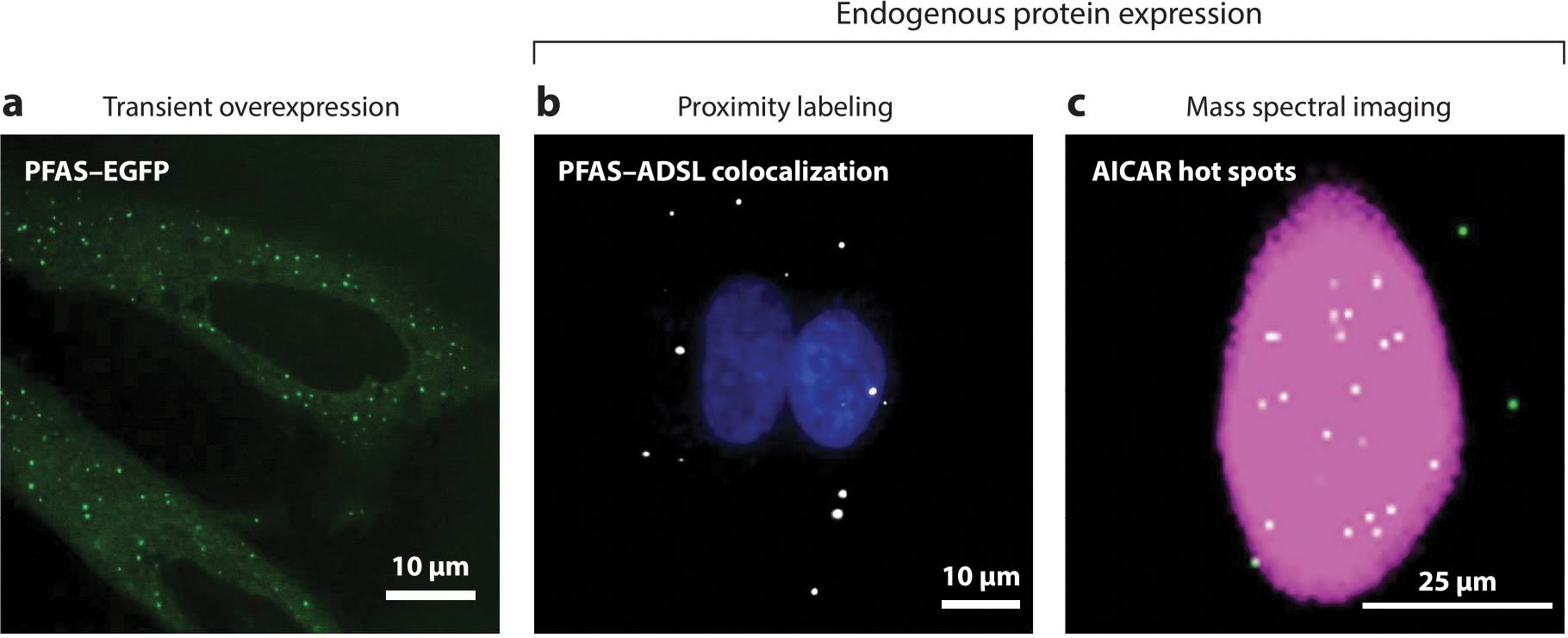
Imaging methods for purinosome detection. Purinosomes can be visualized by three different imaging methods. (a) Initial studies used the transient overexpression of DNPB enzymes tagged with a fluorescent protein such as PFAS–EGFP in purine-depleted HeLa cells to characterize these enzyme assemblies (green punctate staining) (64). The misidentification of purinosomes due to the possibility of DNPB enzyme self-assembly and aggregation resulted in the development of new methods for detecting the complex using endogenous protein expression. These methods include proximity labeling assays (PLAs) and mass spectral imaging. (b) PLAs detect purinosomes through the colocalization of two DNPB enzymes, such as PFAS and ADSL (white) in purine-depleted HeLa cells (17). Nuclei are shown in blue. (c) A representative GCIB-SIMS image of a HeLa cell showing the total ion current (pink) and the pixels with high AICAR abundance (white) (33). These AICAR hot spots also show high levels of ATP, arising as a result of purinosome activity. Abbreviations: AICAR, 5-aminoimidazole-4-carboxamide ribonucleotide; ADSL, adenylosuccinate lyase; DNPB, de novo purine biosynthesis; EGFP, enhanced green fluorescent protein; GCIB-SIMS, gas cluster ion beam–time of flight–secondary ion mass spectrometry; PFAS, phosphoribosyl formylglycinamide synthase; PLA, proximity labeling assay.
