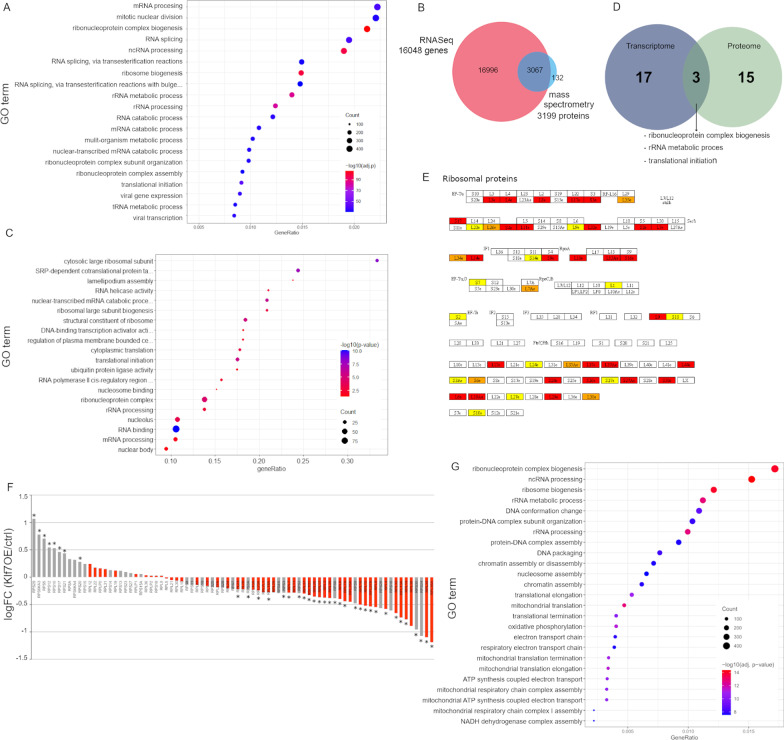
Fig. 2.
Transcriptomic and proteomic data indicated that KLF7 regulated ribosomal pathways. A GSEA of RNA-Seq data showing the top identified GO terms. B Overlap of identified genes and proteins in RNA-Seq and mass spectrometry. C GO term analysis of shot-gun proteomic data depicted the most regulated pathways. D Top significant GO-terms identified in transcriptomics and proteomics data overlapped. E Overlapping KEGG pathway of proteomics and transcriptomics data. Red: significantly altered proteins found in mass spectrometry; yellow: significantly modified genes identified in RNA-Seq; orange: compounds found in both screens. F Expression levels of ribosomal proteins in MS data in KLF7 compared to control. Red: 40S subunit; grey: 60S subunit. *p < 0.05. G Top regulated terms of the gene set enrichment analysis of The Cancer Genome Atlas (TCGA) data in patients stratified according to their KLF7 expression level