Figure 4.
Structural features of BA.2.75 spike RBD and NTD
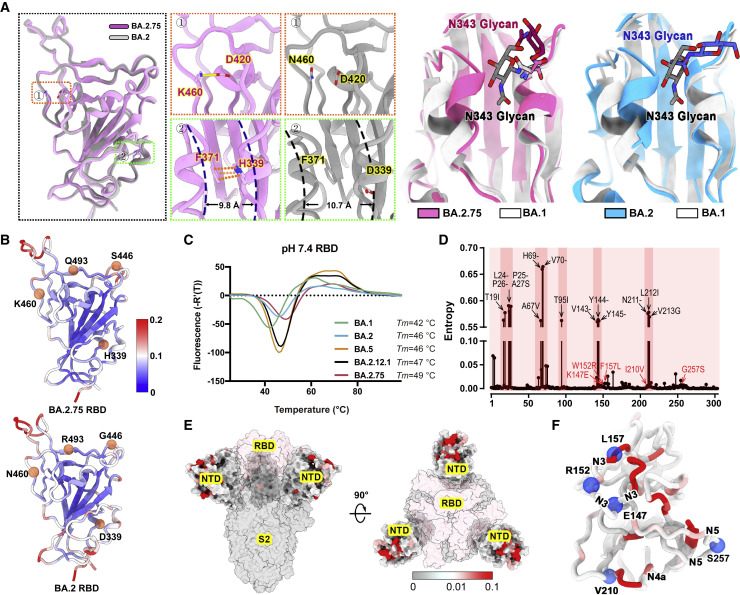
(A) Structural comparisons of RBDs of BA.2.75, BA.2, and BA.1. The newly established interaction of BA.2.75 (pink) with respect to BA.2 (gray) on the RBD is shown on the left. Salt bridges formed between D420 and K460 and π-π stack formed between H339 and F371 in BA.2.75 RBD are highlighted. The distances between α1 and α2 helices on RBD are also marked. A diagram presentation of N343 glycan conformational differences among BA.1 (gray), BA.2 (blue), and BA.2.75 (pink) is shown on the right.
(B) The stability landscapes of BA.2.75 and BA.2 RBD. The cartoons of BA.2.75 and BA.2 RBD are colored by root mean square fluctuation (RMSF) calculated from the last 2 ns of the MD stimulations. Residues 339, 446, 460, and 493 are shown as red spheres.
(C) Thermal stability measurements of the RBD from BA.1 (green), BA.2 (blue), BA.5 (orange), BA.2.12.1 (black), and BA.2.75 (red) at pH 7.4.
(D) Entropy of SARS-CoV-2 NTD variants among circulating isolates. The residues with higher entropy are highlighted by dark red background. The dominant mutations on SARS-CoV-2 NTD and the mutations on BA.2.75 NTD are labeled in black and red, respectively.
(E) The heatmap for circulating variants with mutations on the NTD. Mutation frequency for each residue is calculated based on the datasets from Global Initiative on Sharing All Influenza Data (GISAID).
(F) Cartoon representation of NTD colored by mutation frequency as same as (E). Five mutations in the BA.2.75 NTD are displayed as blue balls and labeled. The secondary structure these mutations locate are also labeled.
See also Figure S3.