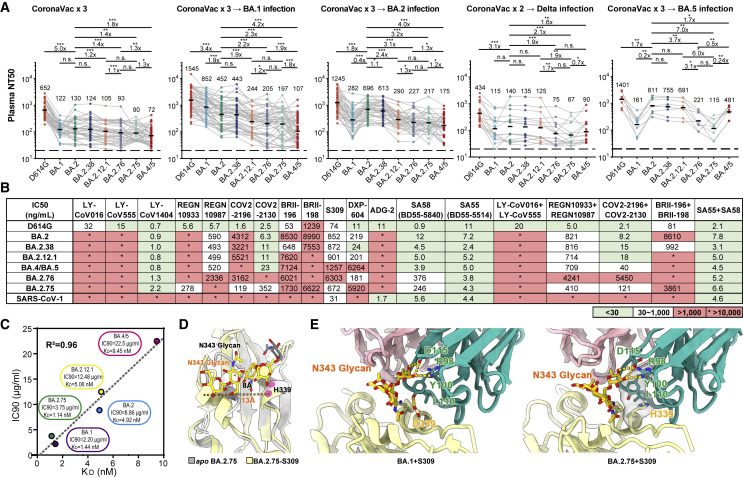
Figure 5.
BA.2.38, BA.2.75, and BA.2.76 showed distinct antibody evasion
(A) Half neutralization titers (NT50s) against SARS-CoV-2 D614G and Omicron variants pseudoviruses by plasma samples from individuals who received 3 doses CoronaVac (n = 40), 3 doses CoronaVac followed by BA.1 infection (n = 50), 3 doses CoronaVac followed by BA.2 infection (n = 39), 2 doses CoronaVac followed by Delta infection (n = 16), or 3 doses CoronaVac followed by BA.5 (n = 8) infection. Geometric mean titers (GMTs) are annotated above each group. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. p values are calculated using two-tailed Wilcoxon signed-rank test of paired samples. Pair-wise fold changes were calculated using the NT50 against the strain on the left divided by that against the strain on the right.
(B) Neutralizing activities against SARS-CoV-2 D614G, Omicron variants, and SARS-CoV-1 pseudovirus of therapeutic neutralizing antibodies. Background colors indicate neutralization levels. Green, IC50 < 30 ng/mL; white, 30 ng/mL < IC50 < 1,000 ng/mL; red, IC50 > 1,000 ng/mL. ∗IC50 > 10,000 ng/mL.
(C) Correlation plots of binding affinities and neutralizing activities (IC90) of S309 against BA.1, BA,2, BA.2.12.1, BA.4/5, and BA.2.75.
(D) Local conformational alterations in the BA.2.75 RBD upon S309 binding. The N343 glycan of the apo BA.2.75 RBD (gray) and S309-bounded BA.2.75 RBD (yellow) are shown as sticks and the distances between α1 and α2 helices from two configurations are also labeled.
(E) Interaction details of the BA.1 RBD (left) and BA.2.75 RBD (right) in complex with S309. Hydrogen bonds and hydrophobic patches are presented as yellow dashed lines and gray surfaces, respectively. The light chain and heavy chain of S309 are colored in pink and cyan, respectively.
See also Figures S4 and S5.