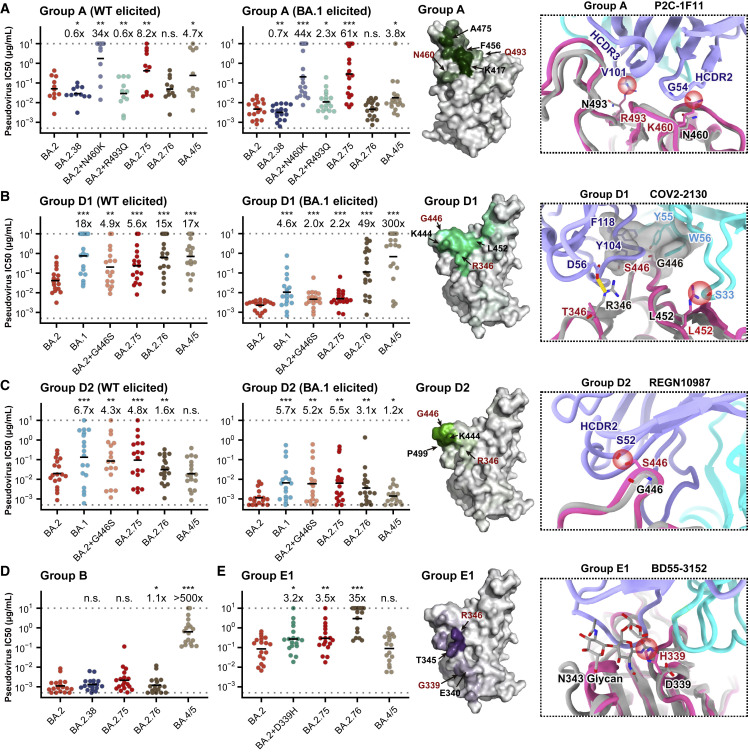
Figure 6.
Evasion of NAbs targeting various RBD epitopes by BA.2.38, BA.2.75, and BA.2.76
Neutralizing activities against SARS-CoV-2 Omicron variants of NAbs in (A) group A (n = 11 isolated from SARS-CoV-2WT convalescents or vaccinees; n = 18 from post-vaccination BA.1 convalescents), (B) group D1 (n = 17, 18, respectively), (C) group D2 (n = 18, 17, respectively), (D) group B (n = 18), and (E) group E1 (n = 18). Deep mutational scanning (DMS) profiles of antibodies in groups A, D1, D2, and E1 are projected onto RBD structure to show interacting hotspots of each group. Color shades indicate escape scores of RBD residues. Interface structural models of representative antibodies in groups A, D1, D2, and E1, in complex of RBD, show potential escaping mechanism of BA.2.75 and BA.2.76. Geometric mean of IC50 fold changes compared with BA.2 are annotated above the bars. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. p values are calculated using two-tailed Wilcoxon signed-rank test of paired samples.