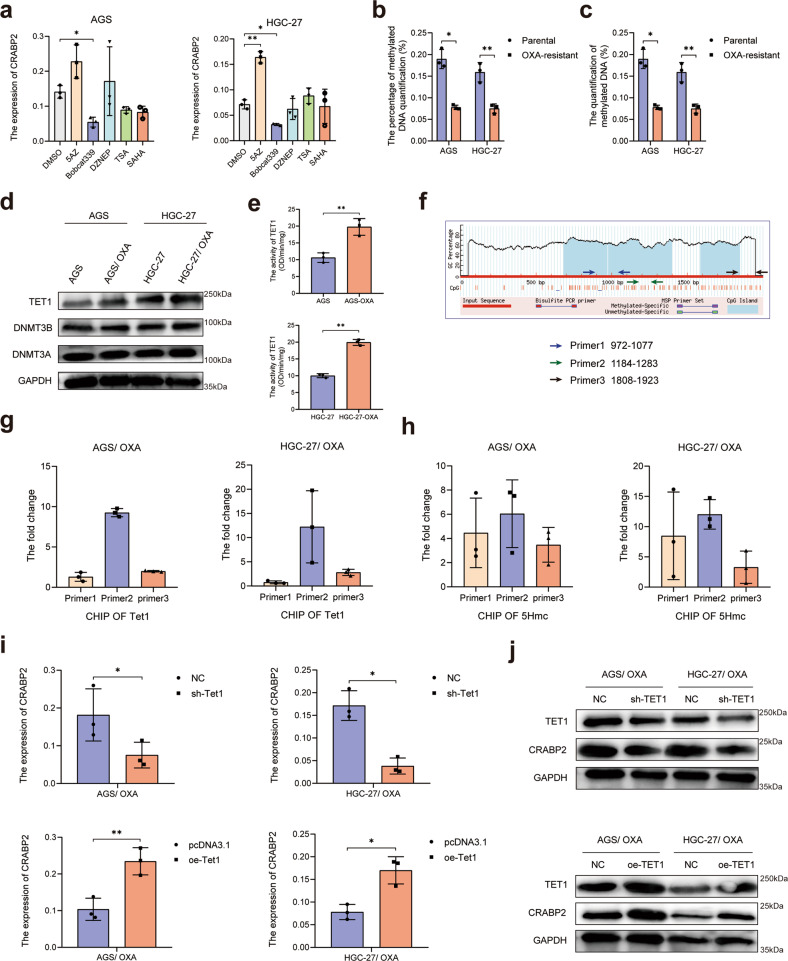
Fig. 6. TET1-mediated DNA hydroxymethylation expedited the expression of CRABP2.
a After adding 5AZ, Bobcat339, DZNEP, TSA, or SAHA to AGS and HGC-27 cells, the mRNA expression changes in CRABP2 were detected. The methylation (b) and hydroxymethylation (c) levels of OXA-resistant GC cells and parental GC cells were examined. d The expression and activity of DNA methylase, demethylase, and hydroxymethylase from OXA-resistant and parental GC cells were examined by western blotting. e The activity of hydroxymethylase TET1 was detected in OXA-resistant GC cells and parental GC cells. f Schematic diagram of the predicted and designed methylation sites in the promoter region of CRABP2. g ChIP experiments using a TET1 antibody were performed in OXA-resistant GC cells. h Hydroxymethylated ChIP assays were performed in OXA-resistant GC cells. After knocking down or overexpressing TET1 in OXA-resistant GC cells, the expression of CRABP2 was examined by qRT–PCR (i) and western blotting (j). *P < 0.05, **P < 0.01, ***P < 0.001, ns not significant.