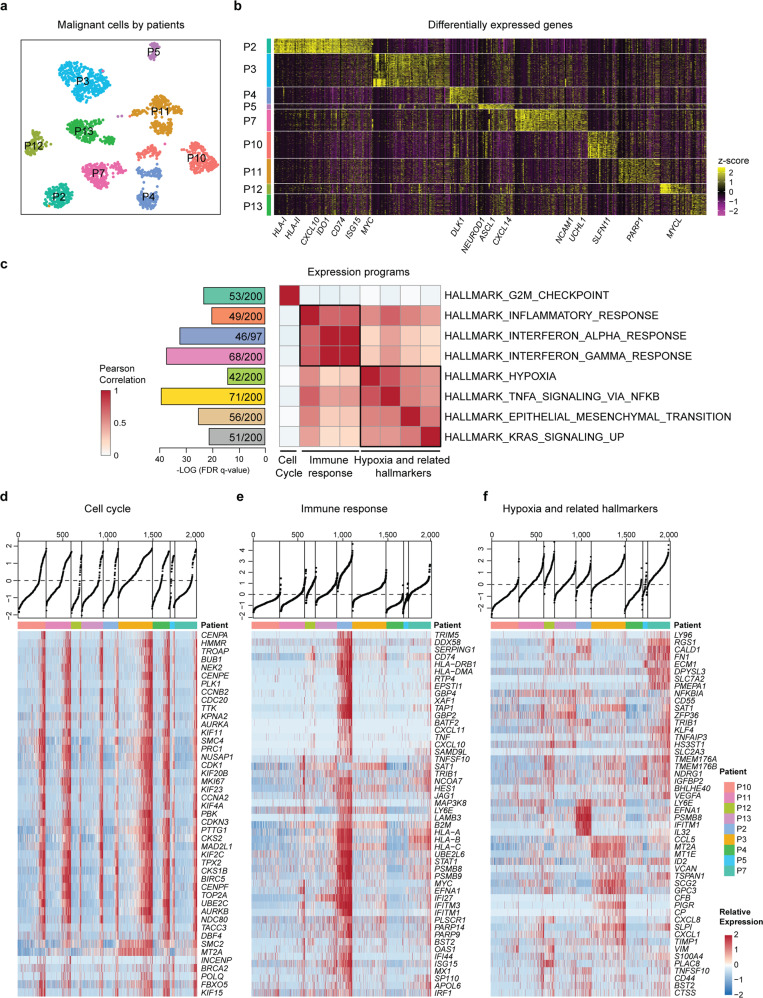
Fig. 3.
Expression heterogeneity of the malignant cell compartments in primary SCLC. a UMAP plot of malignant cells from nine SCLC patients (indicated by color) reveals tumor-specific clusters. b Heatmap of DEGs (top 100 genes in each patient, ranked by the log fold-change in the average expression between cells from one patient and all other cells) across nine individual SCLC primary tumors (columns). Expression data were normalized with z-score transformation, in which yellow and purple represent the high and low expression of a gene, respectively, relative to the median expression level (see the color scale). Selected genes are highlighted. MHC class I molecules (MHC-I) mainly included HLA-A/B/C/E/F; MHC class II molecules (MHC-II) included HLA-DMA/PA1/RA/B1/B5. c GSEA of DEGs (top 2,000 genes ranked by their dispersion values) using hallmark gene set collections. The most enriched hallmarks (ranked by the FDR q-values) are shown with the number of mapped and total genes in the pathways. Pearson correlation analysis was performed based on the mean expression levels of the genes involved in these hallmarks. d Heatmap reveals the expression of the proliferation program and representative genes from the program (rows) in individual SCLC cells (columns). Cells were grouped by tumor and ordered by the single-sample GSEA (ssGSEA) score (top). e Heatmap depicts the expression of the immune program (the inflammatory response, interferon α response, and interferon γ response) as shown in (c) and (f), Heatmap depicts hypoxia-related hallmarks (including TNFα signaling via NF-κB, EMT, KRAS signaling up, and hypoxia) as shown in c