Abstract
Severe acute pancreatitis-associated acute lung injury (SAP-ALI) is a serious disease associated with high mortality. Emodin has been applied to alleviate SAP-ALI; however, the mechanism remains unclear. We report that the therapeutic role of emodin in attenuating SAP-ALI is partly dependent on an exosomal mechanism. SAP rats had increased levels of plasma exosomes with altered protein contents compared to the sham rats. These infused plasma exosomes tended to accumulate in the lungs and promoted the hyper-activation of alveolar macrophages and inflammatory damage. Conversely, emodin treatment decreased the plasma/pancreatic exosome levels in the SAP rats. Emodin-primed exosomes showed less pro-inflammatory effects in alveolar macrophages and lung tissues than SAP exosomes. In detail, emodin-primed exosomes suppressed the NF-κB pathway to reduce the activation of alveolar macrophage and ameliorate lung inflammation by regulating PPARγ pathway, while these effects were amplified/abolished by PPARγ agonist/antagonist. Blockage of pancreatic acinar cell exosome biogenesis also exhibited suppression of alveolar macrophage activation and reduction of lung inflammation. This study suggests a vital role of exosomes in participating inflammation-associated organ-injury, and indicates emodin can attenuate SAP-ALI by reducing the pancreatic exosome-mediated alveolar macrophage activation.
KEY WORDS: Severe acute pancreatitis, Acute lung injury, Emodin, Exosome, Macrophages, Lisocitrate dehydrogenase 1, Peroxisome proliferator-activated receptor γ, Nuclear factor κB
Graphical abstract
Emodin attenuates acute pancreatitis-associated lung injury by reducing pancreatic exosome production and altering exosomal protein contents, which consequently polarizes M2 phenotype of alveolar macrophages via regulating PPARγ-NF-κB pathways.
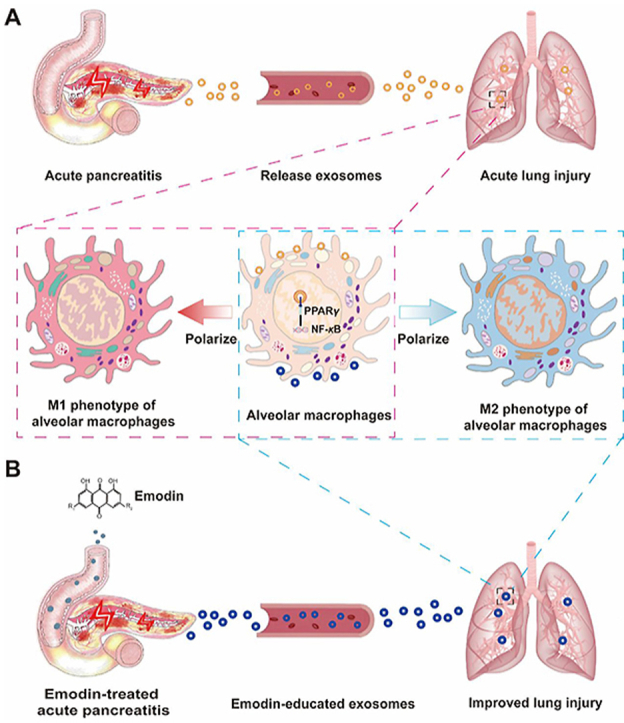
1. Introduction
Acute pancreatitis, is an inflammatory disease of the pancreas in which activated pancreatic enzymes attack the pancreas, resulting in a pancreatic dysfunction, followed by local and systemic inflammations1,2. Approximately 20%–30% of the patients with acute pancreatitis exhibit progression to severe acute pancreatitis (SAP), which is characterized by local complications or distant organ dysfunctions and is associated with high morbidity and mortality rates3. Lung injury is the most common extra pancreatic organ dysfunction induced by SAP4, 5, 6, 7, and approximately one in three patients with SAP will develop acute lung injury (ALI) or even the acute respiratory distress syndrome (ARDS)8,9. The mortality rate of AP with complications of ALI is 30%–40%10,11. SAP-associated lung injury (ALI) is characterized by an increased permeability of the pulmonary microvasculature and leakage of protein-rich exudate into the alveolar space, followed by alveolar congestion and interstitial edema9,12. Non-invasive/invasive ventilation, antibiotics, and specific diets have been used to suppress lung inflammation clinically13; however, the mechanisms underlying the development of SAP-ALI remain elusive and effective interventions to attenuate SAP-ALI are still lacking. Therefore, it is important to explore the potential mechanisms underlying SAP-ALI development and to investigate therapeutic drugs that might be effective against this condition.
Emodin (1,3,8-trihydroxy-6-methylanthraquinone), a natural anthraquinone derivative found in various Chinese medicinal herbs, such as Rheum palmatum, Polygonum cuspidatum, Polygonum multiflorum, Aloe vera, and Cassia obtusifolia14, exhibits multiple beneficial effects, including antibacterial, anti-inflammatory, and anti-hyperlipidemic effects15. In preclinical SAP models, emodin has been reported to alleviate SAP and SAP-ALI by modulating the lung inflammatory microenvironment16,17. In rat models of SAP, emodin attenuated oxidative stress and inflammation by inhibiting NF-κB signaling18,19. In addition, emodin has been found to inhibit NLRP3 inflammasome activation20, promote neutrophil apoptosis21, and inhibit neutrophil protease activity in the lungs of SAP-ALI rats22.
Hyper-activated alveolar macrophages have been recognized as key mediators of SAP-ALI, as they are the main source of local pro-inflammatory cytokines (e.g., TNF-α), inflammatory substances (e.g., NO), and chemokines (e.g., MIP-2) and are capable of amplifying lung inflammation by recruiting circulating neutrophils11,23, 24, 25. Conversely, the re-polarization or depletion of pro-inflammatory alveolar macrophages has been reported to attenuate lung inflammation in SAP-ALI to some extent12,23,26. Nonetheless, the effect of emodin on alveolar macrophage activation during SAP-ALI remains unclear.
Increasing evidence suggests the existence of exosome-based communication between SAP and ALI. Exosomes are small (∼30–150 nm) extracellular vesicles released by living cells that play important roles in intercellular communication by transferring information (nucleic acids, proteins, and lipids) from host cells to neighboring or distant target cells27 and thus are involved in mediating various pathological processes28. After AP, the exosome levels in ascitic fluid and the blood circulation are markedly increased, and the increased plasma exosome levels are likely to be the consequence of pancreatic damage and are partly responsible for AP-associated lung inflammation29. In ALI model mice, dye-labeled plasma exosomes infused via the tail vein reached the lungs, where they were internalized by alveolar macrophages30. A further study revealed that the serum exosomes delivered miR-155 to alveolar macrophages, thereby activating nuclear factor (NF)-κB signaling and promoting local inflammation in the lungs of mice with sepsis31. Plasma exosomes induced lung injury by activating NLRP3-dependent pyroptosis of alveolar macrophages in l-arginine-induced AP mice32. Altogether, these findings suggest that circulating exosomes in SAP may be involved in mediating lung inflammation and ALI33; however, the possible effect of emodin on these phenomena during SAP-ALI is unclear.
In this study, we hypothesized that emodin may attenuate SAP-ALI by preventing pancreatic exosome-induced lung inflammation. To investigate this hypothesis, we established a rat model of SAP-ALI and evaluated the changes in lung lesions, pancreatic and plasma exosomes, and macrophage activation after treatment with emodin. The effect of emodin-educated exosomes on the phenotype of alveolar macrophages was evaluated in vitro and in vivo. In addition, exosomal contents and the possible mechanism of emodin-induced changes were analyzed using proteomic analyses, RNAseq and network pharmacology.
2. Materials and methods
2.1. Antibodies and reagents
Sodium taurocholate and emodin were obtained from Beijing Zhongke Quality Inspection Biology Technology Company (Beijing, China). GW9662 (MB5023), GW4869 (MB0764), and rosiglitazone (MB1211) were provided by Dalian Meilun Biotechnology (Dalian, China). Anti-CD68 (MA5-28262) and anti-CD163 (PA5-78961) were purchased from Thermo Fisher Scientific (MA, USA), anti-CD86 (551396) was purchased from BD Biosciences (San Diego, CA, USA), anti-PPARγ (sc-7273) was procured from Santa Cruz Biotechnology (Santa Cruz, CA, USA), phospho-PPAR gamma (Ser112) antibody (AF3284) and β-actin antibody (AF7018) were obtained from Affinity Biosciences (OH, USA), anti-NF-κB P65 (phosphor S529, ab97726), anti-CD63 (ab134045), anti-TSG101 (ab125011), and anti-Calnexin (ab22595) were purchased from Abcam (Milton, UK), anti-IDH1 (GB111194) was purchased from Servicebio (Wuhan, China), anti-RAB27A (#69295) and NF-κB p65 (#8242) were obtained from Cell Signaling Technology (Boston, USA), and anti-GAPDH (10494-1-AP) was purchased from Proteintech Company (Chicago, USA). The anti-CD68 (ab201340) and anti-TNF-α (NBP1-19532) used for the immunohistochemistry were procured from Abcam and Novus Biological, respectively. A rat tumor necrosis factor α (TNF-α) ELISA kit was obtained from Multi Sciences (LIANKE) Biotech (Hangzhou, China). The rat interleukin 10 (IL-10) ELISA Kit (E-EL-R0016c) was purchased from Elabscience Biotechnology Co., Ltd. (Wuhan, China). A rat macrophage inflammatory protein-2 (MIP-2) ELISA kit, nitric oxide (NO) determination kit, and myeloperoxidase (MPO) assay kit were purchased from Jiancheng (Nanjing, China). The fluorescent dye for the exosomal DIR labeling (UR21017) was purchased from Umibio (Shanghai, China).
2.2. Animals
Male SD rats (250–300 g) were obtained from Chengdu Dashuo Laboratory Animal (Chengdu, China). The rats were housed in a controlled environment and had ad libitum access to standard laboratory pelleted formula and tap water. All the animal experiments conformed with the Guidelines for the Care and Use of Laboratory Animals issued by the National Academy of Sciences, published by the National Institute of Health. The experiments were approved by the Experimental Animal Ethics Committee of the Sichuan University of West China Hospital (Approval Number: 2020247A).
2.3. Rat SAP model establishment and treatment
A rat model of taurocholate-induced AP was established as described previously34. Briefly, rats were anesthetized using an intraperitoneal injection of 10% chloral hydrate (0.3 mL/100 g body weight). The biliopancreatic duct was cannulated through the duodenum and a small bulldog clamp was used to close the hepatic duct. AP was induced using a retrograde perfusion of 3.5% sodium taurocholate at a constant speed of 6 mL/h with a perfusion pump (LD-P2020II, Lande Medical Appliance Co., Ltd., Shanghai, China). The control animals (sham group) were treated with the same volume of normal saline (0.9% NaCl). After the injection, the upper and lower ends of the biliopancreatic duct were blocked for 5 min. Then, the microvascular clip was removed, the puncture hole sutured with silk thread, and the rats placed back in their cages after two layers of abdominal closure. Sixteen SAP rats were randomly divided into the emodin treatment group and the SAP group according to their numbers (the treatment group with an odd number and the SAP group with an even number, and with eight rats in each group) due to that the 24-h survival rate of SAP rats was 80%–90%. The rats in the emodin treatment group were provided 10 mg/kg emodin per gavage 12 or 18 h after the induction of AP, whereas the rats in the SAP alone group and sham group (eight rats subjected to laparotomy) were provided the same amount of normal saline. The operators who established the SAP model were blinded to the treatment, i.e., they did not know which SAP rats belonged to the SAP group or the emodin treatment group. Twenty-four hours after treatment, blood and pancreatic tissues were collected and processed immediately to isolate the exosomes. Bronchoalveolar lavage fluid (BALF) was collected to analyze the macrophage phenotype analysis and an in-vitro culture. All the experiments were independently performed three times. In addition, the pancreas and lung samples were harvested, frozen, and stored at −80 °C until further analysis.
2.4. Isolation of the rat primary acinar cells
After removing the fat tissues and blood vessels, the fresh pancreatic tissues were washed twice with HEPES (pH = 7.0) without calcium and magnesium and cut into 1-cubic-millimeter fragments using a small pair of scissors. The fragments were digested using 10 mL collagenase type I (200 U/mL, Gibco) by gently shaking on a shaker at 37 °C for 20 min. The DEME-F12 (Servicebio, Wuhan, China) medium containing 20% serum was added to terminate the reaction. The lysates were collected and filtered through a 70-μm filter (BD Falcon, USA). After centrifugation at 500 ×g for 5 min, lysis for 1 min, and two washes, the isolated pancreatic acinar cells were resuspended in the DEME-F12 medium containing 20% serum (Gibco) and 1% penicillin-streptomycin and cultured at 37 °C in a humidified chamber containing an atmosphere of 5% CO2.
2.5. Isolation of exosomes
Fresh rat pancreatic tissue was cut into 1-cubic-millimeter fragments and digested using collagenase type I (200 U/mL) at 37 °C for 20 min to isolate the pancreatic exosomes. After centrifugation at 2000 ×g, 4 °C for 10 min, the supernatants were collected and used for exosome extraction. For the isolation of plasma exosomes, rat plasma samples were centrifuged at 1900 ×g, 4 °C for 15 min, and the top layer of plasma was collected and used for the exosome extraction. For the isolation of cellular exosomes, the culture medium was collected and used for the exosome extraction.
Exosomes were isolated from the tissue lysates, plasma, and culture medium using a modified differential ultracentrifugation protocol (Beckman Coulter, CA, USA). In the first step, the samples were centrifuged at 2000 ×g, 4 °C for 20 min to remove large cell fragments or debris. The supernatants were collected and centrifuged at 12,000 ×g, 4 °C for 45 min to remove small cellular debris. Then, the supernatants were transferred to a new tube and ultracentrifuged at 120,000 ×g, 4 °C for 120 min to pellet the small vesicles. The supernatants were discarded, and the pellets were resuspended in a large volume of PBS, and then filtered through a 0.22-μm filter to eliminate potential contaminants. After ultracentrifugation at 120,000 ×g, 4 °C for 120 min, the pellets were resuspended in PBS, and the exosomal protein concentrations were determined using a bicinchoninic acid (BCA) assay kit (P0010S, Beyotime Biotechnology, Shanghai, China).
2.6. Isolation of the alveolar macrophages
After anesthesia, the tracheae of the rats were exposed and intubated with a 14G intravenous indwelling needle (Shanghai Puyi Medical Equipment Co., Ltd.). Lung lavage fluid (10 mL, sterilized saline) was collected using a 10-mL syringe (Shanghai Kindly Medical Instruments). After centrifugation at 250 ×g for 5 min, the supernatant of the lung lavage fluid was used to detect the presence of inflammatory mediators, while the pelleted cells were washed with PBS and used for flow-cytometric analysis of the macrophage phenotype. For culturing alveolar macrophages, the lavage fluid was centrifuged at 180 ×g for 5 min and re-suspended in RPMI-1640 medium containing 10% fetal calf serum, penicillin (100 U/mL), and streptomycin (100 μg/mL). After incubation for 2 h at 37 °C in an atmosphere of 5% CO2, non-adherent cells were removed.
2.7. Tracking of circulating exosomes in vivo
Plasma exosomes were labeled with 10 μmol/L DIR dye (Ex/Em: 748 nm/780 nm) for 30 min, and then centrifuged at 100,000 ×g for 90 min to remove the excess dye. The exosomes were washed with sterile PBS and then centrifuged at 100,000 ×g for 90 min. The stained exosomes were resuspended in 100 μL of sterile PBS and the protein concentrations were determined using the BCA assay. For the in-vivo analysis of the exosome distribution, DIR-labeled exosomes (100 μg protein) were administered via the tail vein into rats (80–100 g). Twenty-four hours after injection, the rats were euthanized using an intraperitoneal injection of 10% chloral hydrate (0.3 mL/100 g body weight) and the organs were harvested and imaged using an IVIS spectrum in vivo imaging system (IVIS Lumina II, PerkinElmer, MA, USA). The fluorescence intensity in the images was analyzed using the IVIS software (Ex/Em: 748 nm/780 nm, Living Image Software for IVIS).
2.8. Cellular exosome uptake analysis
Plasma exosomes were labeled with DIR (5 μmol/L) at room temperature for 10 min and then washed with sterile PBS, followed by centrifugation at 100,000 ×g for 90 min. The stained exosomes were resuspended in 40 μL of sterile PBS. Alveolar macrophages were incubated with 20 μg/mL exosomes for 24 h. After the incubation, the cells were washed three times with PBS and fixed with 4% paraformaldehyde at room temperature for 30 min. After staining with Hoechst (Invitrogen, #33342) for 10 min, the sections were sealed and observed under a confocal microscope (N-SIM S, Nikon).
2.9. Serum enzymes and inflammatory-related factors assays
The levels of amylase and lipase in the rat sera were measured using standard assays on an automatic biochemistry analyzer (West China-Frontier PharmaTech, Chengdu, China). The concentrations of TNF-α, MIP-2, NO, and IL-10 in rat BALF, cell culture medium supernatant and lung tissue lysate were analyzed according to the manufacturer's instructions. The concentration of MPO in the lung tissues was also measured according to the manufacturer's instructions.
2.10. Exosome characterization
The isolated exosomes were characterized using transmission electron microscopy (TEM; HT-7700, Hitachi, Japan) and nanoparticle tracking analysis (NTA, NanoFCM, N30E). Briefly, exosomes were fixed in 2% paraformaldehyde, adsorbed on formvar-coated nickel grids, and stained negatively with 4% uranyl oxalate. The grids were air-dried and observed by TEM. The particle size and the concentration of the exosomes were evaluated in accordance with instructions provided along with the nanoparticle tracking analyzer. The exosomes were subjected to Western blotting with specific antibodies against exosomal marker proteins (CD63 and TSG101) and a negative marker (CALNEXIN).
2.11. Proteomic analysis of plasma exosomes
Total protein was extracted from the plasma exosome samples; one part was used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) after the determination of the concentration, while the other part was digested with trypsin. Tryptic peptide mixtures labeled with tandem mass tags (Thermo Scientific, Rockfors, IL, USA) were prepared according to the standard procedure. Six labeled peptide mixtures were combined and subjected to reversed-phase chromatographic separation and mass spectrometry analysis by Shanghai Luming Biological Technology. Database searching was conducted using Proteome Discover 2.4 (Thermo Fisher, San Jose, CA, USA) with the following parameters: static modification: tandem mass tag 6-plex (N-term, K); dynamic modification: oxidation (M), acetyl (N-term); digestion: trypsin; ms1 tolerance: 10 ppm; ms2 tolerance: 0.02 Da; missed cleavages: 2. Gene Ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were used for biological categorization of the significantly differentially expressed proteins under SAP conditions.
2.12. Network pharmacology analysis
Target genes of emodin were first screened using TCMSP (http://tcmspw.com/index.php), DrugBank (https://go.drugbank.com/drugs), chEMBL (https://www.ebi.ac.uk/chembl), PubChem (http://pubchem.ncbi.nlm.nih.gov), and the therapeutic Target Database (db.idrblab.net/ttd/) data. The emodin target genes and differentially expressed proteins in the plasma exosomes in SAP conditions were imported into String (http://string-db.org/, ver.10) to establish a potential protein-protein interaction (PPI) network33. Cytoscape (http://cytoscape.org/, ver3.7.2) was used to visualize the network34.
2.13. RNA sequencing
Primary acinar cells were isolated from the pancreatic tissues of healthy rats and cultured in vitro. They were divided randomly into the normal (NG) group, NaTC group, and NaTC + Emodin group. The NG group did not receive any treatment. The NaTC group was stimulated with 500 μmol/L sodium taurocholate for 1 h, while the NaTC + Emodin group was treated with 20 μmol/L emodin for 0.5 h, and then stimulated with 500 μmol/L sodium taurocholate for 1 h. Total RNA was extracted from the acinar cells using the TRIzol Reagent according the manufacturer's instructions (Invitrogen, USA) and genomic DNA was removed from the preparation using DNase I (TaKara, Japan). Total RNA was extracted from three biological replicates. RNA quality was assessed using the 2100 Bioanalyser (Agilent, USA). Library preparation and sequencing was performed by Majorbio Biotech (Shanghai, China) on an Illumina HiSeq xten/NovaSeq 6000 sequencer. The data were analyzed on the free online Majorbio I-Sanger Cloud Platform (www.i-sanger.com).
2.14. Flow-cytometric analysis of alveolar macrophages
After centrifugation at 100 ×g, 4 °C for 7 min, pelleted cells from the lavage fluid were re-suspended in 100 μL of staining buffer containing 6 μL anti-CD163 (M2 marker) and incubated in the dark at 4 °C for 30 min. After two washes, 1 μL of IgG (AF647) was added to 100 μL of the resuspended cells (about 5 × 105 cells), which were then incubated in the dark at 4 °C for 1 h. Pelleted cells were washed three times and then stained with 100 μL of buffer containing 5 μL FITC-labeled anti-CD68 (M1 marker) and 5 μL of PE-labeled anti-CD86 in the dark at 4 °C for 30 min. The stained cells were resuspended in 100 μL of the wash buffer and analyzed using flow cytometry (Cytoflex, Beckman).
2.15. Western blotting
The tissues or cells were lysed in an appropriate amount of RIPA buffer and centrifuged at 12,000 ×g for 5 min. The supernatants were collected and protein concentrations determined using a BCA kit. A 12% SDS-PAGE gel was used to obtain a high-resolution separation of the protein samples (20 or 40 μg). These proteins were transferred onto nitrocellulose membranes or polyvinylidene difluoride membranes (Millipore, USA). The membranes were blocked using a commercial blocking solution (EpiZyme Biotech, Shanghai, China) at room temperature for 10 min, washed three times with TBST, and incubated with the primary antibodies against TSG101 (1:1000, Abcam), CD63 (1:1000, Abcam), Calnexin (1:1000, Abcam), p-PPARγ (1:500, Affinity), PPARγ (1:500, Santa), p-NF-κB (1:800, Abcam), NF-κB (1:1000, CST), GAPDH (1:20000, Proteintech), andβ-actin (1:20000, Affinity) at 4 °C, overnight. The next day (after 12 h), the membranes were washed three times with TBST and then incubated with the appropriate horseradish peroxidase (HRP)-labeled secondary antibody for 60 min. After three washes, the blots were visualized using an imaging system (Bio-Rad, CA, USA).
2.16. Histopathological examination
The collected rat pancreatic and lung tissues were fixed with 4% formaldehyde. The paraffin-embedded tissues were cut into 4-μm sections, dewaxed twice using xylene, and washed with ethanol to remove the xylene. The tissue sections were stained with hematoxylin for 5 min and eosin for 2 min. Images of the stained sections were acquired using a Pannoramic Digital Slide Scanners (3D HISTECH, Hungary). As previously described35, pancreatic tissue sections were scored for the severity of pancreatitis based on edema (0–4 points), inflammation, vacuolization (0–4 points), and necrosis (0–4 points). The individual scores were then added to obtain the total pathological score for the pancreatic tissue. The degree of pulmonary injury was assessed using a scale for the interstitial and intra-alveolar edema, interstitial and intra-alveolar leukocyte infiltration, and fibrosis, as previously described36. Each item was scored using a 5-point scale as follows: 0) minimal damage, 1) mild damage, 2) moderate damage, 3) severe damage, and 4) maximal damage. The sum of the scores was the pathological score of the lung tissue.
2.17. Immunohistochemistry
The paraffin-embedded pancreatic and lung tissues were sectioned at 4 μm and mounted on APES-coated slides (AR0001, Boster Biological Technology Co., Ltd., Wuhan, China). The sections were deparaffinized using xylene and rehydrated in graded ethanol. The endogenous peroxidase activity was quenched with 3% hydrogen peroxide at room temperature for 30 min, followed by rinsing in PBS. After antigen retrieval in a 10 mmol/L citrate buffer (pH = 6.0) at 94 °C for 10 min, the sections were incubated with blocking solution at room temperature for 1 h to block non-specific site. The sections were then incubated with primary antibodies against TNF-α (1:200, Novus) and CD68 (1:400, Abcam) at 4 °C overnight (about 12 h). After three washes, the sections were incubated with the appropriate secondary antibodies at room temperature for 30 min, followed by incubation with streptavidin-HRP reagent and DAB substrate. The sections were then immersed in deionized water and counterstained with hematoxylin, washed, and sealed.
2.18. Statistical analysis
All data showed a normal distribution, and they are expressed as mean ± standard deviation (SD) and analyzed using GraphPad Prism 8 (GraphPad Software, CA, USA). The significance was evaluated using an unpaired two-tailed Student's t-test (two groups) or one-way analysis of variance (ANOVA) followed by Tukey's multiple comparisons tests (multiple groups). P < 0.05 was considered to be significant.
3. Results
3.1. Emodin has a therapeutic effect on SAP-ALI
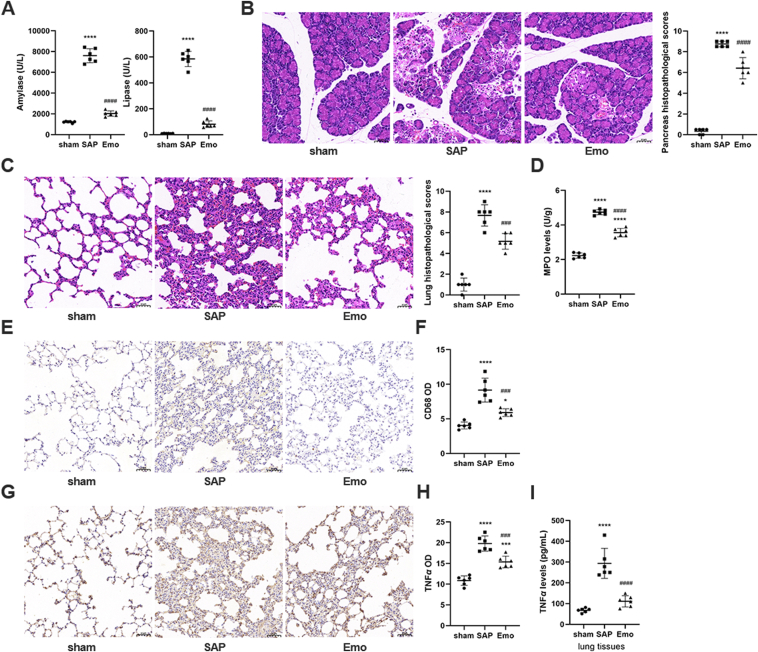
Compared with sham rats, the serum amylase and lipase levels in SAP rats were increased (P < 0.0001), and necrosis, inflammation, and hemorrhage, were observed in the pancreas at six hours after SAP induction (Supporting Information Fig. S1A and S1B). The pancreatic pathological scores also increased gradually after SAP induction (Fig. S1C). However, there was no significant difference in the lung pathological scores between the sham and SAP groups 6 h after SAP induction, and only slight pathological thickening of the lung tissue was observed in rats of the SAP group (Fig. S1B and S1D). Severe pulmonary damage appeared 12 h after SAP induction, as indicated by the alveolar wall thickening, bleeding, and neutrophil infiltration, as well as the increase in the lung injury score. Therefore, emodin was administrated to the rats at 12 h (i.e., the time point at which ALI occurred) after SAP induction. As no significant differences were observed between the sham + emodin group and sham group with respect to the pathology of the pancreas and lung tissues (Supporting Information Fig. S2), the sham group was used as the control group in the subsequent experiments. And the high dose of emodin (10 mg/kg) was found to be more effective than the low dose of emodin (5 mg/kg) at decreasing lipase and amylase levels and lowering the pathological scores for the pancreas and lung tissues (Supporting Information Fig. S3). Thus, 10 mg/kg of emodin was selected in the following experiments. As shown in Fig. 1A–C, the emodin significantly treatment decreased the amylase, lipase, pancreatic and lung injury pathological score in SAP rats (P < 0.0001; compared with those in the SAP group). Moreover, rats in the SAP group showed higher CD68+ macrophage counts and pro-inflammatory factor (MPO and TNF-α) levels in lung tissues than those in the sham group, whereas the levels of these inflammatory markers in lung tissues were markedly reduced after the emodin treatment (Fig. 1D–J).
Figure 1.
Emodin has a therapeutic effect on SAP-ALI. (A) Serum amylase and lipase levels in the sham operation group (sham), SAP model group (SAP), and SAP plus emodin treatment group (Emo). (B) Morphological observation of pancreas using H&E staining (scale bar = 50 μm) and pancreatic histopathological scores. (C) Morphological observation of the lungs by H&E staining (scale bar = 50 μm) and lung histopathological scores. (D) MPO levels in lung tissues of rats (n = 6). (E) CD68 expression in the lung tissues of rats evaluated using immunohistochemistry. (F) Quantification of CD68 expression in the lung tissues (n = 6). (G) TNF-α expression in the lung tissues of rats evaluated using immunohistochemistry. (H) Quantification of TNF-α expression in the lung tissues (n = 6). (I) TNF-α levels evaluated by ELISA in lung tissues of rats. All experiments were performed three times, independently. All data were expressed as mean ± SD. ∗P < 0.05, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001 vs. the sham group; ###P < 0.001 and ####P < 0.0001 vs. the SAP group, by one-way ANOVA followed by Tukey tests.
3.2. Characterization of plasma exosomes in SAP-ALI
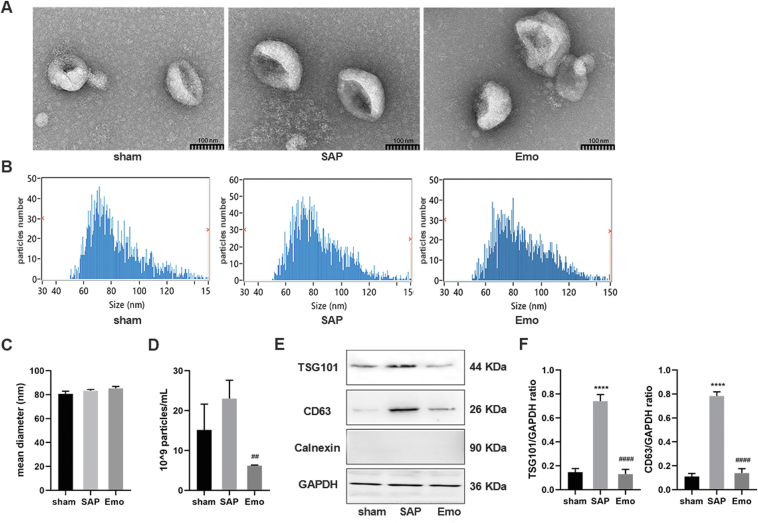
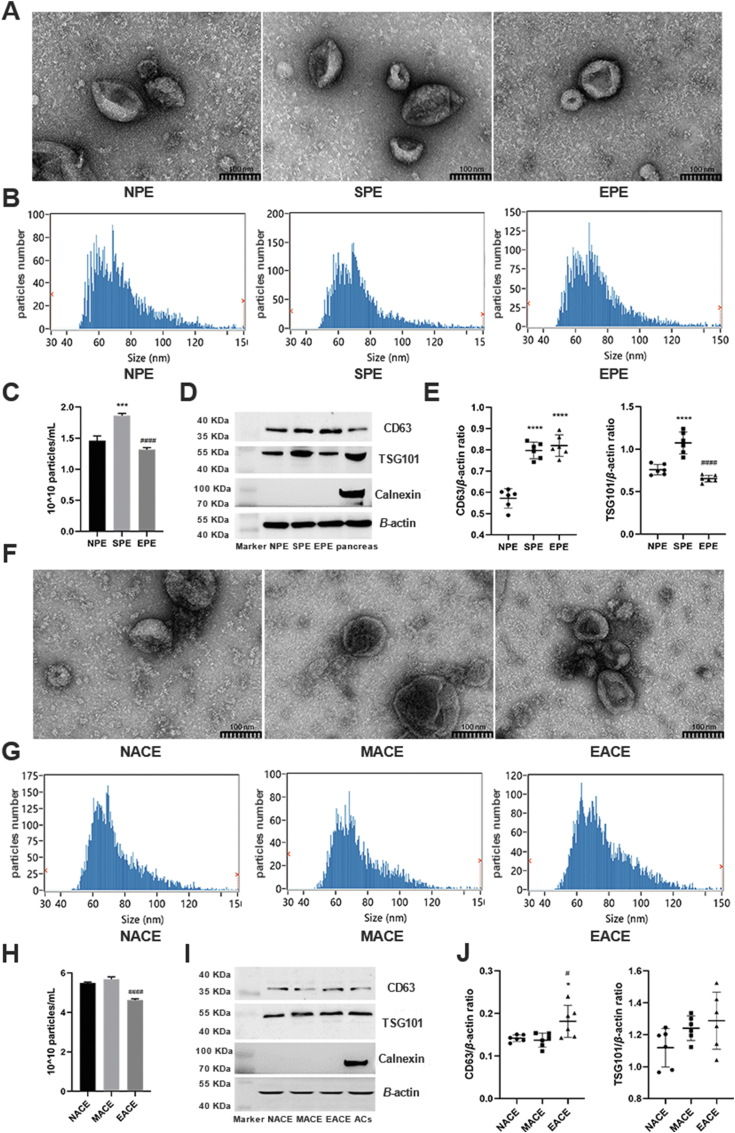
To explore the changes in plasma exosomes during SAP-ALI and to investigate whether they were affected by emodin, the plasma exosomes isolated from rats of each group were analyzed. The TEM and NTA revealed that plasma exosomes in the three groups were typically cup-shaped, with sizes of 60–120 nm (Fig. 2A and B). The average exosome size in the emodin-treated group was showed a slight increased (∼86 nm); however, there was no significant difference between the groups (P > 0.05) (Fig. 2C). The SAP rats exhibited a higher number of exosomes in the plasma, which was reduced after treatment with emodin (P < 0.01, Fig. 2D). In all three groups, the plasma exosomes expressed the positive markers (TSG101 and CD63), but not the negative marker (Calnexin, Fig. 2E). The levels of TSG101 and CD63 were increased in plasma exosomes from SAP rats, but were suppressed by the emodin treatment (Fig. 2E and F).
Figure 2.
Characterize of plasma exosomes in SAP-ALI. (A) Representative TEM images of plasma exosomes in rats of each group, scale bar = 100 nm. (B) Size distribution of plasma exosomes in each group as detected by NTA. (C) Plasma exosome sizes as detected by NTA (n = 3). (D) Plasma exosome concentrations (n = 3). (E) Western blot analysis of TSG101, CD63, and Calnexin levels in plasma exosomes. GAPDH was used as the loading control. (F) Quantitative analysis of TSG101 and CD63 expression in plasma exosomes (n = 3). All experiments were performed three times, independently. ∗∗∗∗P < 0.0001 vs. the sham group; ##P < 0.01 and ####P < 0.0001 vs. the SAP group, by one-way ANOVA followed by Tukey tests.
3.3. Emodin ameliorates SAP exosome-induced lung inflammation in vivo
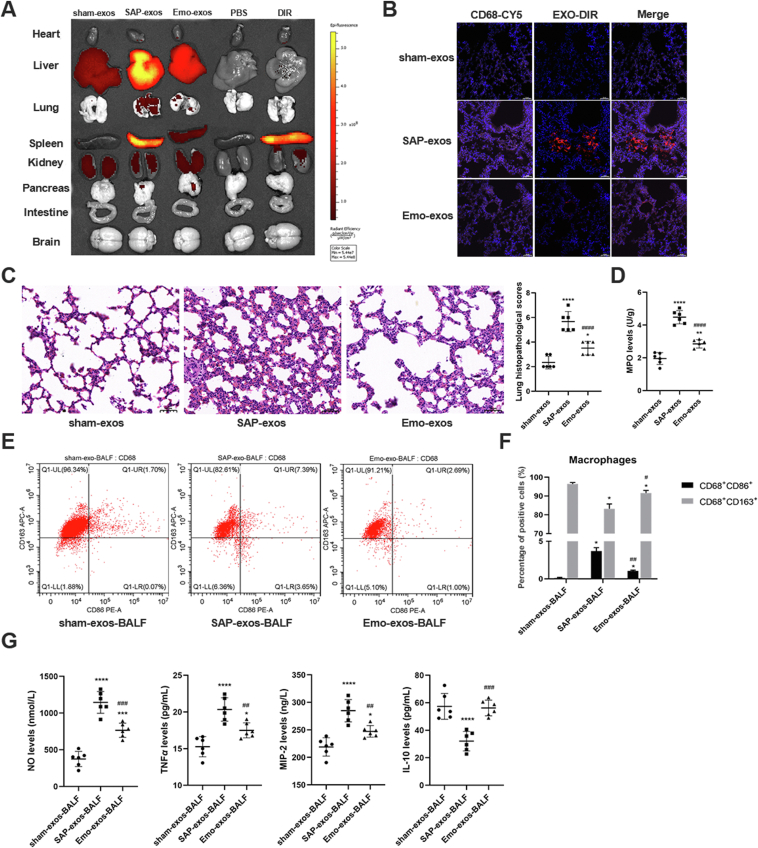
To investigate the effect of emodin on the uptake of circulating exosomes in vivo, the plasma exosomes isolated from rats from the different treatment groups were labeled with the DIR dye and then injected into healthy rats via the tail vein. As shown in Fig. 3A, the infused plasma exosomes were mainly taken up by the liver, lungs, spleen, and kidneys. Exosomes from the SAP rats (SAP-exos; accordingly, sham-exos and emo-exos indicate exosomes from rats in the sham- and emodin-treated groups, respectively) displayed higher accumulation in the liver, spleen, lungs, and pancreas than those in the sham- or emo-exos groups; exosome accumulation in these tissues was higher than that in the heart and small intestine. H&E staining revealed that lung was the most severely damaged organ upon exosome treatment (Fig. 3C and Supporting Information Fig. S4). The DIR-labeled plasma exosomes were observed in the lung tissue sections, and some of them co-localized with the CD68+ macrophages (Fig. 3B).
Figure 3.
Emodin ameliorates SAP exosome-induced lung inflammation in vivo. (A) Exosomes of the three groups (sham-exos, SAP-exos, and Emo-exos) were labeled with DIR in vitro, and fluorescence images of isolated organs were obtained at 24 h after intravenous injection of the DIR-labeled exosomes into healthy rats. (B) Representative fluorescence images of lung tissue sections from the three groups. CD68 was labeled with CY5, emitting pink fluorescence, and exosomes emitted red fluorescence, scale bar = 50 μm. (C) Morphological observation of lung tissues and lung histopathological scores (n = 6), scale bar = 50 μm. (D) MPO expression in the lung tissues of rats (n = 6). (E, F) Twenty-four hours after tail-vein injection of exosomes, BALF was collected and analyzed for the expression of macrophage marker CD68, M1 macrophage marker CD86, and M2 macrophage marker CD163 using flow cytometry. (G) Levels of the pro-inflammatory factors NO, TNF-α, MIP-2, and anti-inflammatory factor IL-10 in BALF as detected by ELISA (n = 6). All experiments were performed three times, independently. All data were expressed as mean ± SD. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001 vs. the sham-exos group; #P < 0.05, ##P < 0.01, ###P < 0.001, and ####P < 0.0001 vs. the SAP-exos group, by one-way ANOVA followed by Tukey tests.
Compared with the rats that had been administered sham-exos, the rats that had been administered SAP-exos had increased lung injury scores and expressions of MPO in the lung tissues, whereas the degree of lung inflammatory injuries was lower in the rats treated with emo-exos than that in the rats treated with SAP-exos (Fig. 3C and D). The BALF was collected from rats and analyzed 24 h after the exosome injection. Compared with those in the sham-exos group, the BALF from the rats in the SAP-exos group showed higher M1 macrophage counts (CD68+/CD86+) and pro-inflammatory factor (NO, TNF-α, and MIP-2) levels, but lower M2 macrophage (CD68+/CD163+) counts and anti-inflammatory factor levels (IL-10) (P < 0.05, and P < 0.01, respectively). Conversely, the rats in the emo-exos group had lower M1 macrophage counts and pro-inflammatory factor levels, as well as higher M2 macrophage counts and anti-inflammatory factor levels than those in the SAP-exos group (P < 0.05 and P < 0.001, respectively, Fig. 3E–G).
3.4. Emodin suppresses SAP exosome-induced alveolar macrophage activation in vitro
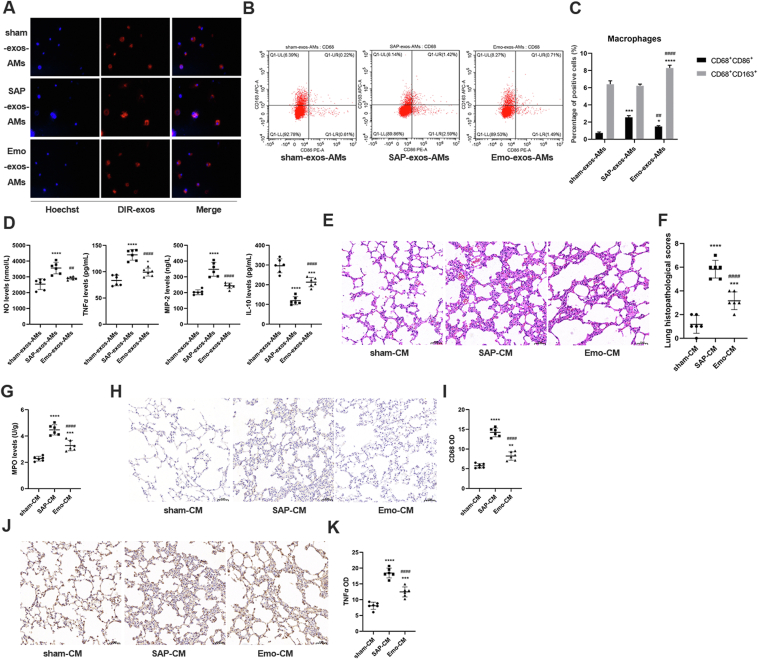
As emodin was found to reduce SAP-exos-induced lung injury in vivo, we next explored the impact of circulating exosomes on the phenotypes of alveolar macrophages in vitro. As shown in Fig. 4A, the plasma exosomes from the rats in the different groups were taken up effectively by the alveolar macrophages in vitro. Compared with those in the sham-exos group, the alveolar macrophages in the SAP-exos group expressed higher levels of the M1 macrophages marker CD68+CD86+ (P < 0.0001), whereas the macrophages exposed to emodin-exos expressed lower levels of the M1 marker CD68+CD86+ and higher levels of the M2 markers CD68+CD163+ than macrophages in the SAP-exos group (P < 0.01, P < 0.001, respectively, Fig. 4B and C). Next, we analyzed the changes in the inflammatory factor levels in the culture medium of alveolar macrophages. The levels of NO, TNF-α, and MIP-2 were increased in the culture medium of alveolar macrophages exposed to SAP-exos (P < 0.001), but were decreased in response to the emo-exos treatment (P < 0.01). In contrast, IL-10 levels were decreased in the SAP-exos-intervened culture medium compared to that of the alveolar macrophages exposed to sham-exos, whereas the emo-exos increased the IL-10 levels in the culture medium (P < 0.001) (Fig. 4D). Conditioned media from alveolar macrophage cultures was injected into the lungs of healthy rats via tracheal administration to investigate the pathogenic effects. As shown in Fig. 4E–K, the conditioned medium from the SAP-exos group induced alveolar wall thickening and neutrophil infiltration, and increased the levels of the MPO, CD68, and TNF-α in lung tissues (P < 0.0001), whereas treatment with the conditioned medium from the emo-exos group resulted in lower lung injury scores and MPO, CD68, and TNF-α levels than those in the SAP-exos group (P < 0.001, P < 0.0001, respectively).
Figure 4.
Emodin suppresses SAP exosome-induced alveolar macrophages activation in vitro. (A) Plasma exosomes were labeled with DIR and incubated in vitro with primary alveolar macrophages for 24 h. Representative images acquired by confocal microscopy are shown. (B) Flow cytometric analysis of M1 (CD86+) and M2 (CD163+) markers in macrophages (CD68+) incubated with the exosomes. (C) Quantitative results of flow cytometry. (D) Levels of the pro-inflammatory factors NO, TNF-α, MIP-2, and anti-inflammatory factor IL-10 in culture medium as detected by ELISA (n = 6). (E) Morphological observation of lung tissues of healthy rats after treatment with conditioned medium (CM) of macrophages, scale bar = 50 μm. (F) Lung histopathological scores corresponding to panel E (n = 6). (G) MPO expression in the lung tissues of rats (n = 6). (H) CD68 expression in the lung tissues of rats as detected by immunohistochemistry, scale bar = 50 μm. (I) Quantitative results of CD68 expression in the lung tissues (n = 6). (J) TNF-α expression in the lung tissues of rats as detected by immunohistochemistry, scale bar = 50 μm. (K) Quantitative results of TNF-α expression in the lung tissues (n = 6). All experiments were performed three times, independently. All data were expressed as mean ± SD. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001 vs. the sham-CM group; ##P < 0.01, and ####P < 0.0001 vs. the SAP-CM group, by one-way ANOVA followed by Tukey tests.
3.5. Emodin suppresses pancreatic exosome secretion during SAP
To explore the effect of emodin on pancreatic exosome production in SAP conditions, the exosomes secreted from pancreatic tissues and acinar cells were isolated and analyzed. The results showed that the exosomes secreted by the pancreatic tissues exhibited a characteristic cup shape with sizes of 50–100 nm (Fig. 5A and B). In line with the changes in the plasma exosomes, the exosome secretion from the pancreases of the SAP rats (SPE, accordingly NPE and EPE, indicate exosomes from the pancreases of sham rats and emodin-treated SAP rats, respectively) increased significantly (P < 0.001) when compared to that of the sham group, whereas emodin treatment reduced the SAP-related pancreatic exosome secretion (Fig. 5C, P < 0.0001). Exosomes from the pancreas expressed the positive markers TSG101 and CD63, but not the negative marker CALNEXIN (Fig. 5D). Next, we analyzed the in-vitro exosome production in rat pancreatic acinar cells treated with sodium taurocholate or emodin. The exosomes secreted by the acinar cells were cup-shaped—small vesicles of 50–100 nm in size—and expressed TSG101 and CD63, but not Calnexin (Fig. 5F–J). The number of exosomes released by the acinar cells treated with sodium taurocholate (MACE, accordingly NACE and EACE, indicate exosomes from the acinar cells of PBS and emodin-treated, respectively) was slightly higher than that in the control group (P > 0.05), whereas emodin suppressed the exosome production in these cells under conditions of sodium taurocholate stimulation (Fig. 5H, P < 0.0001).
Figure 5.
Emodin suppresses pancreatic exosome secretion during SAP. (A) Representative TEM images of exosomes secreted by the pancreas in the sham operation group (NPE), SAP model group (SPE), and emodin treatment group (EPE). (B) Size distribution of pancreatic exosomes in each group, scale bar = 100 nm. (C) Quantitative analysis of pancreatic exosome concentration (n = 6). (D) Western blot analysis of TSG101, CD63, and Calnexin in pancreatic exosomes. β-actin was used as the loading control, and pancreas lysate was used as the control. (E) Quantitative analysis of CD63 and TSG101 expression in pancreatic exosomes (n = 6). (F) Representative TEM pictures of exosomes secreted by acinar cells in the NACE group treated with PBS, MACE group (treatment with 500 μmol/L sodium taurocholate for 1 h), and EACE group (pretreatment with 20 μmol/L emodin for 30 min, followed by treatment with 500 μmol/L sodium taurocholate for 1 h), scale bar = 100 nm. (G) Size distribution of exosomes secreted by acinar cells in each group. (H) Quantitative analysis of exosome production in acinar cells (n = 6). (I) Western blot analysis of TSG101, CD63, and Calnexin in exosomes secreted by acinar cells. β-actin was used as the loading control, and acinar cells lysate was used as the control. (J) Quantitative analysis of CD63 and TSG101 expression in exosomes secreted by acinar cells (n = 6). All experiments were performed three times, independently. ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.001 vs. the NPE or NACE group; #P < 0.05, ####P < 0.0001 vs. the SPE or MACE group, by one-way ANOVA followed by Tukey tests.
To further explore the effect of emodin on exosome-derived maternal pancreatic acinar cells, RNA-Seq was used to analyze emodin-induced gene expression in the acinar cells damaged by sodium taurocholate (NaTC). The results reveal 90 genes in acinar cells may be affected by emodin under NaTC condition (Supporting Information Fig. S5 and Table S1). Further GO and KEGG annotation and enrichment analyses revealed that emodin could affect multiple pathways in acinar cells, such as the those involving localization, immune responses, response to external stimuli, multiple infectious diseases, signal interaction, and NF-κB pathways (Supporting Information Figs. S5–S7). To evaluate the effect of emodin on exosome secretion of pancreatic acinar cells, we examined the expression of RAB27A in the pancreas and acinar cells. The results show that both the pancreatic tissue of the rats in the SAP group and acinar cells treated with NaTC expressed higher levels of RAB27A, while emodin treatment suppressed the upregulation of RAB27A in damaged pancreas and acinar cells (Supporting Information Fig. S8A and S8B).
3.6. Effect of emodin on plasma exosomal protein contents
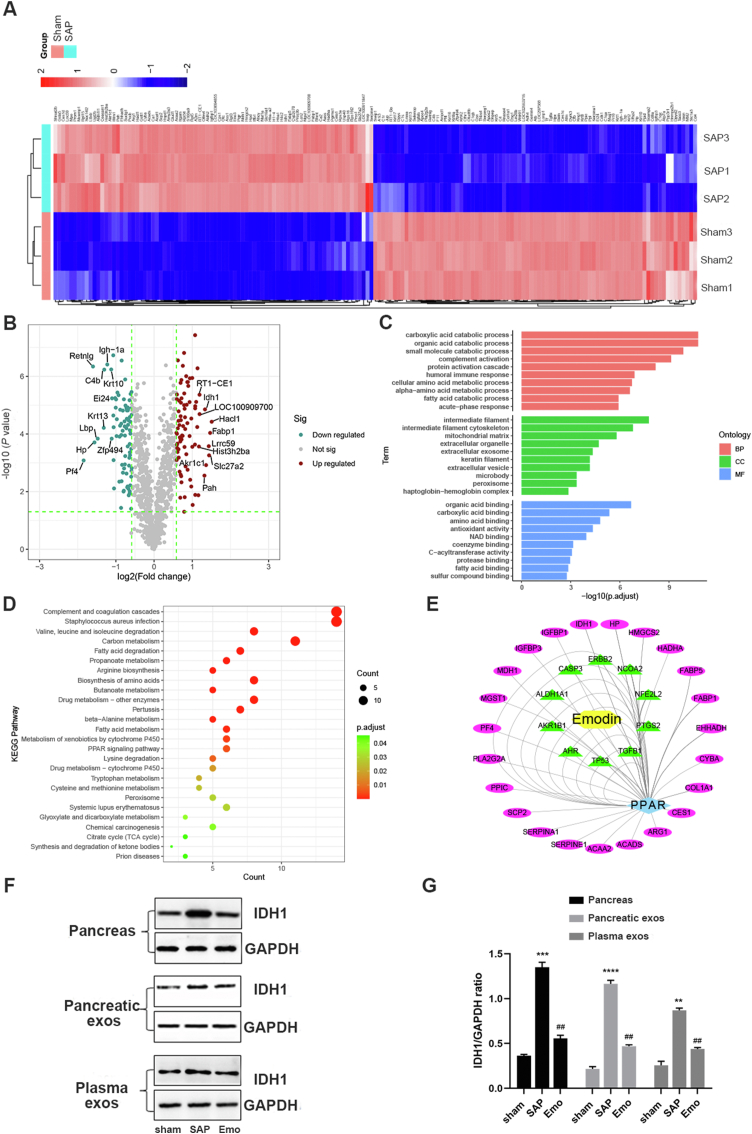
Next, we analyzed changes in the protein contents of the plasma exosomes in SAP rats using proteomic analyses. The heatmap in Fig. 6A shows 165 significantly differentially expressed proteins (|logFC| >0.58; SAP group versus sham group). Among these, 82 proteins were up-regulated and 83 proteins were down-regulated (Supporting Information Table S2). The volcano plot in Fig. 6B shows the top 10 proteins in the exosomes (SAP group versus sham group), including HACL1, FABP1, SLC27A2, LRRC59, AKR1C1, IDH1, PAH, LOC100909700, RT1-CE1, and HIST3H2BA. The differentially expressed proteins were subjected to GO term (biological process, cell component, and molecular function) and KEGG pathway enrichment analyses to reveal their biological functions (Fig. 6C and D). The exosomal proteins expressed differentially in the SAP group were involved in multiple processes, including the degradation of organic acids and the binding of the mitochondrial matrix to organic acids (Supporting Information Tables S3–S8), as well as carbon metabolism and peroxisome proliferator-activated receptor γ (PPARγ) signaling (Fig. 6D). Network pharmacology analysis was performed to explore the possible interactions between emodin and these differentially expressed exosomal proteins. PPARγ signaling was identified as a major pathway affected by emodin via indirect downstream proteins, such as IDH1, FABP1, HMGCS2, and SCP2E (Fig. 6E). The changes in the expression of several potential downstream proteins of emodin were validated by Western blotting. For example, the IDH1 levels were higher in the pancreatic tissues, pancreatic exosomes, and plasma exosomes of SAP rats than in those of sham rats, whereas the emodin treatment reduced the SAP-induced IDH1 levels in exosomes and pancreatic tissues (Fig. 6F and G).
Figure 6.
Effect of emodin on plasma exosomal protein contents. (A) Heatmap analysis of exosome proteins in the sham operation group and SAP group (n = 3). (B) Volcano plot of differentially expressed proteins between the sham operation and SAP groups. (C, D) GO term and KEGG pathway analyses of differentially expressed exosomal proteins between the sham operation and SAP groups. (E) Network pharmacology analysis of possible target pathways of emodin and plasma exosomal proteins in the sham and SAP groups. (F) Western blot analysis of IDH1 in pancreatic tissues, pancreatic exosomes, and plasma exosomes of the sham, SAP, and emodin groups. GAPDH was used as the loading control (G) Quantitative analysis of IDH1 expression (n = 3). ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.001 vs. the sham group; ##P < 0.01 vs. the SAP group, by one-way ANOVA followed by Tukey tests.
3.7. Effect of emodin on the PPARγ-NF-κB pathway in macrophages and lung tissues
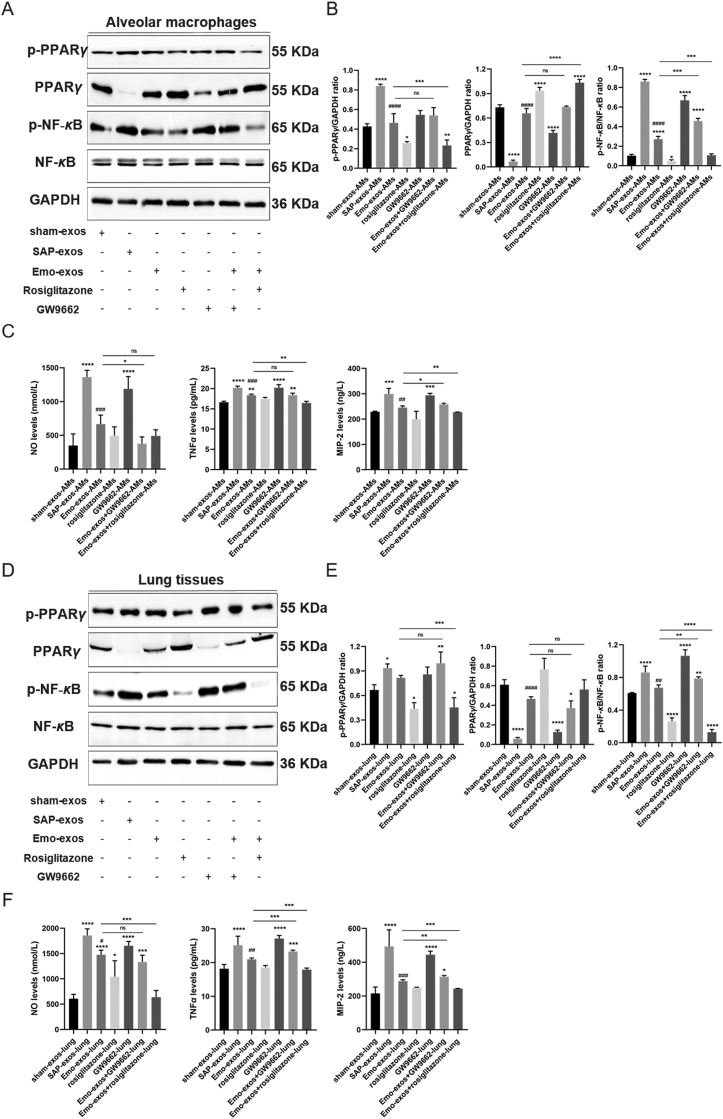
Next, we evaluated the effects of different exosomes on PPARγ and its downstream NF-κB pathway in alveolar macrophages and lung tissues. Compared with sham-exos, SAP-exos reduced the level of PPARγ and increased that of p-PPARγ and p-NF-κB in alveolar macrophages and lung tissues, whereas the cells and lung tissues in the emodin-exos group had higher levels of PPARγ and lower levels of p-PPARγ and p-NF-κB than those in SAP-exos group (Fig. 7A, B, D and E). Consistent with previous reports37, 38, 39, 40, 41, rosiglitazone inhibited PPARγ phosphorylation at the S112 residue, and partly upregulated the expression of total PPARγ in macrophages and downregulated the expression of p-NF-κB. GW9662 had no effect on the expression of p-PPARγ, but decreased the expression of total PPARγ and increased the expression of p-NF-κB. Moreover, GW9662 did not affect the regulation of p-PPARγ and PPARγ expression by emodin-exos in alveolar macrophages, while rosiglitazone enhanced this regulation. Furthermore, rosiglitazone reduced the expression of p-NF-κB in macrophages treated with emodin-exos, whereas GW9662 exerted an opposite effect, i.e., increased the p-NF-κB levels reduced by emodin (Fig. 7B, P < 0.001). Thus, the effects of emodin-exos in promoting PPARγ expression and suppressing p-NF-κB expression in macrophages were amplified upon co-treatment with rosiglitazone. Macrophages treated with SAP-exos exhibited high expression of the pro-inflammatory mediators (NO, TNF-α, and MIP-2) than those in the sham-exos group, whereas the expression of these pro-inflammatory factors was lower in macrophages treated with emodin-exos than that in macrophages treated with SAP-exos (Fig. 7C).
Figure 7.
Effect of emodin on PPARγ-NF-κB pathway in macrophages and lung tissues. (A) Western blot analysis of p-PPARγ, PPARγ, p-NF-κB, and NF-κB in alveolar macrophages exposed to plasma exosomes from sham rats, SAP rats, and emodin-treated SAP rats and subsequently treated with the PPARγ agonist rosiglitazone or the antagonist GW9662. GAPDH was used as the loading control. (B) Quantitative analysis of p-PPARγ, PPARγ, and p-NF-κB expression in alveolar macrophages (n = 3). (C) Levels of the pro-inflammatory factors NO, TNF-α, and MIP-2 in culture media as detected by ELISA (n = 3). (D) Western blot analysis of p-PPARγ, PPARγ, p-NF-κB, and NF-κB in the lung tissues of healthy rats administered plasma exosomes from sham rats, SAP rats, and emodin-treated SAP rats and subsequently treated with the PPARγ agonist rosiglitazone or the antagonist GW9662. GAPDH was used as the loading control. (E) Quantitative analysis of p-PPARγ, PPARγ, and p-NF-κB expression in the lung tissues (n = 3). (F) Levels of the pro-inflammatory factors NO, TNF-α, and MIP-2 in the lung tissues as detected by ELISA (n = 3). All experiments were independently performed three times. All data were expressed as mean ± SD. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001 vs. the sham group, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001 vs. the Emo-exos-AMs or Emo-exos-lung; #P < 0.05, ##P < 0.01, ###P < 0.001 and ####P < 0.0001 vs. the SAP group, by one-way ANOVA followed by Tukey tests.
In the lung tissues, rosiglitazone decreased the expression of p-PPARγ and p-NF-κB, but had no effect on the expression of total PPARγ. In contrast, GW9662 decreased the expression of total PPARγ in the lung tissue and increased the expression of p-NF-κB, while having no effect on p-PPARγ expression. Rosiglitazone decreased the expression of p-PPARγ and p-NF-κB in the lung tissue in the background of emodin-exos treatment, whereas the inhibitory effect of emodin-exos on p-NF-κB expression was abolished by GW9662. Moreover, emodin-exos decreased the expression of pro-inflammatory factors (NO, TNF-α, and MIP-2), which are known to be induced in response to SAP-exos treatment. Activation of PPARγ by rosiglitazone had no effect on the expression of the pro-inflammatory factors in alveolar macrophages and lung tissues, while the inhibition of PPARγ by GW9662 resulted in increased expression of NO, TNF-α, and MIP-2 in alveolar macrophages and rat lung tissues (Fig. 7C and F).
3.8. Effect of emodin-primed exosomes on alveolar macrophages and SAP-ALI rats
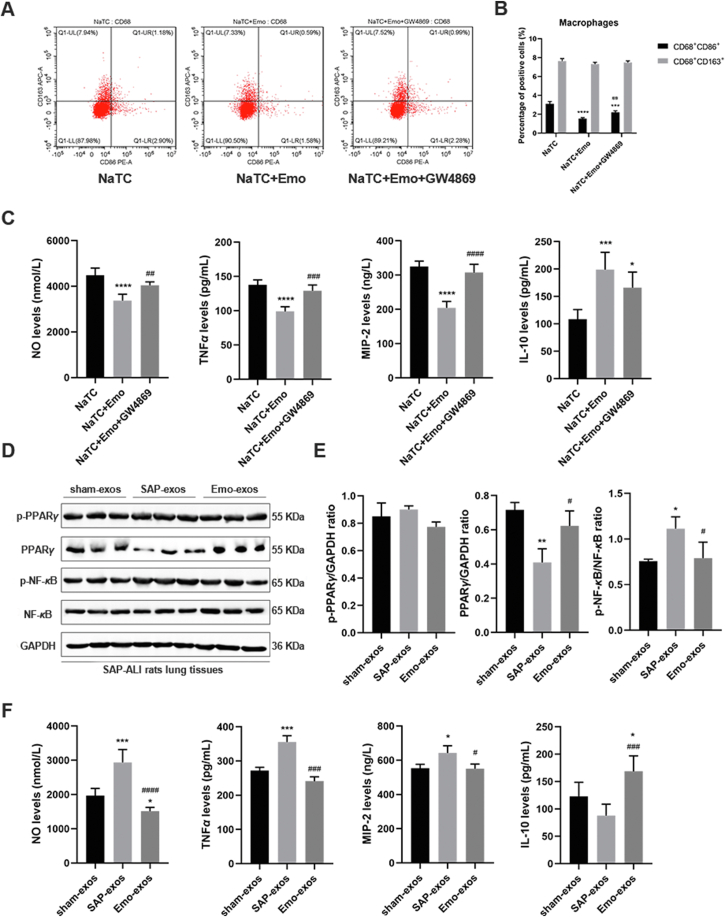
As emodin ameliorated SAP-ALI at least partly by influencing the pancreatic exosome-induced effect, we tried to block the exosomes production demonstrate the role of the exosomes in mediating the effects of emodin. First, we stimulated the pancreatic acinar cells with NaTC to simulate the state of AP, and then used GW4869 (an exosome biogenesis inhibitor) to block the exosomes production in vitro. After that, the supernatants of the pancreatic acinar cells containing the exosomes (in different conditions) were collected to treat the alveolar macrophages. The changes in alveolar macrophage polarization and pro-inflammatory or anti-inflammatory factor release were analyzed. As shown in Fig. 8A and B, compared with the supernatant from the NaTC alone group, the supernatant from the NaTC + emodin group significantly decreased the proportion of the M1 alveolar macrophages (CD68+CD86+), the expression of pro-inflammatory factors (NO, TNF-α, and MIP-2), and increased the expression of anti-inflammatory factors (IL-10) in these macrophages (P < 0.001, Fig. 8C). However, when the exosomes production of acinar cells was blocked using GW4869 in advance, the proportion of the M1 alveolar macrophages increased significantly (P < 0.01), accompanied by increased expression of pro-inflammatory factors compared to those in the NaTC + emodin group.
Figure 8.
Effect of emodin-primed exosomes on alveolar macrophages and SAP-ALI rats. (A) Flow cytometric analysis of M1 (CD86+) and M2 (CD163+) markers in primary rat alveolar macrophages (CD68+) incubated with the conditioned medium (CM) obtained from acinar cells treated with 500 μmol/L sodium taurocholate (treatment for 1 h, NaTC), 20 μmol/L emodin (pretreatment for 30 min, Emo) or 10 μmol/L GW4869 (pretreatment for 30 min). (B) Quantitative results of flow cytometry. (C) Levels of the pro-inflammatory factors NO, TNF-α, MIP-2, and anti-inflammatory factor IL-10 in culture medium of alveolar macrophages as detected by ELISA (n = 6). (D) Western blot analysis of p-PPARγ, PPARγ, p-NF-κB, and NF-κB in the lung tissues of SAP-ALI rats administered plasma exosomes from sham rats, SAP rats, and emodin-treated SAP rats. GAPDH was used as the loading control. (E) Quantitative analysis of p-PPARγ, PPARγ, and p-NF-κB expression in the lung tissues (n = 3). (F) Levels of pro-inflammatory (NO, TNF-α, MIP-2) and anti-inflammatory (IL-10) factors in the lung tissues as detected by ELISA (n = 3). All experiments were performed independently three times. All data were expressed as mean ± SD. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001 vs. the sham group, #P < 0.05, ##P < 0.01, ###P < 0.001 and ####P < 0.0001 vs. the SAP group, by one-way ANOVA followed by Tukey tests.
Next, to validate the beneficial role of the emodin-exos in the lung tissues of SAP-ALI rats in vivo, three groups of plasma exosomes (sham-exos, SAP-exos and Emo-exos) were injected into the SAP-ALI rats via their tail veins. As shown in Fig. 8D and E, the three groups of exosomes did not affect the expression of p-PPARγ in the lung tissues of the SAP-ALI rats. Compared with sham-exos, the SAP-exos groups exhibited decreased expression of PPARγ and increased expression of p-NF-κB in the lung tissues; however, no significant difference was observed in the expression of PPARγ and p-NF-κB between the sham-exos and the emo-exos group. However, emo-exos significantly increased the expression of PPARγ and decreased the expression of p-NF-κB (P < 0.05) in the lung tissues of the SAP-ALI rats (compared to the SAP-exos group). Moreover, compared to sham-exos group, the lung tissues exhibited higher levels of the pro-inflammatory factors (NO, TNF-α, and MIP-2) in the SAP-exos group. In contrast, emo-exos significantly reduced these pro-inflammatory factors levels and increased the expression of IL-10 in the lung tissues of SAP-ALI rats (compared with the SAP-exos group, Fig. 8F).
4. Discussion
While it has been demonstrated that emodin attenuates SAP-ALI21, the detailed protective mechanism has not been completely unraveled. Exosomes are live cell-secreted small vesicles that transfer various host cell-derived bioactive molecules to adjacent or distant cells via the circulatory system28. Circulating exosome levels are increased after SAP, which is thought to be a consequence of pancreatic injury29. In line with these reports, we found higher levels of exosomes in the plasma and pancreatic tissues of SAP rats. Emodin effectively reduced these levels in SAP rats. Moreover, emodin reduced exosome production in pancreatic acinar cells under conditions of sodium taurocholate stimulation. These results suggest that the increased plasma exosome levels in SAP rats can likely be attributed to enhanced exosome secretion from damaged pancreatic acinar cells under SAP conditions. In fact, exosome secretion has been proposed as a means of attenuating cellular stress. Stress or disease states can modulate the amount and composition of exosomes, thereby regulating in vivo immunity42,43. However, the specific mechanisms which enable the sorting of cargos (e.g., protein) into exosomes are complicated; most of which are still not fully understood43. We found that emodin affected the expression of genes related to intracellular protein transport and cellular component organization in damaged acinar cells. In fact, these pathways are known to be involved in regulating various stages of exosome biogenesis, such as formation of MVBs and amphisomes43. For example, it has been reported that amphisome formation and the subsequent exosome release were dependent on ATG5, RAB11, and RAB27A (a key protein that controls the secretion of exosomes)44,45. These findings were partly verified by the detection of RAB27A in pancreatic tissues or in pancreatic acinar cells. The expression of RAB27A in sodium-taurocholate-stimulated pancreatic tissues and acinar cells increased, while emodin treatment in this background resulted in decreased expression of RAB27A. Furthermore, plasma exosomes from SAP rats exhibited a tendency to accumulate in the lungs of rats when compared to those from sham rats, whereas this effect was abrogated by emodin intervention. These results indicate that the increased amounts of pancreatitis-related exosomes in the peripheral circulation of the SAP rats may have been an important factor contributing to acute lung injuries, with emodin being able to at least partly reduce the secretion of harmful pancreatic exosomes in SAP rats, to alleviate SAP-ALI.
It has been recently reported that circulating exosomes in SAP conditions may enter distant organs, such as the lungs, and thus may be involved in the development of lung injuries through the transfer of biological information30. In this study, we also found that the plasma exosomes from the SAP rats accumulated in the lungs, one of the main exosomes gathering organs, and entered the alveolar macrophages, leading to an M1 polarization of the alveolar macrophages and the release of pro-inflammatory mediators, such as NO, TNF-α, and MIP-2 in the lungs. Alveolar macrophages have been considered to be the major mediators of SAP-ALI12, and hyperactivated alveolar macrophages can secrete various pro-inflammatory factors, including cytokines and NO, thereby directly damaging pulmonary cells and indirectly recruiting numerous immune cells (e.g., neutrophils) to aggravate ALI46. Thus, the circulating exosomes—as transmitters of information—may be involved in inducing lung damage under SAP conditions by polarizing pro-inflammatory alveolar macrophages. In contrast, emodin-exos had a lower potential to induce M1 polarization and pro-inflammatory cytokine production in alveolar macrophages and rat lung tissues. These findings suggest that emodin-primed exosomes carry different biological information (e.g., proteins), thereby attenuating lung damage by inhibiting the pro-inflammatory polarization of alveolar macrophages.
Next, we analyzed the changes in the exosomal proteins that may be affected by emodin in SAP-ALI. Through the proteomic analysis of the altered proteins in the plasma exosomes and network pharmacology analysis of the emodin-exosomal protein interactions, PPARγ signaling was identified as a major pathway affected by emodin. PPARγ has been shown to decrease the transcriptional activity of NF-κB and pro-inflammatory polarization of alveolar macrophages47. Furthermore, emodin has been found to regulate the activity of PPARγ-related signaling proteins, such as IDH148, FABP149, and HMGCS250,51 by targeting genes, such as NCOA2, CASP3, AKR1B1, AHR, TGFB1, etc. For example, IDH1 is an isocitrate dehydrogenase found in the cytoplasm and peroxisomes, which is known to be involved in regulating oxidative stress, metabolism, cancer, inflammation, and autophagy52. Importantly, IDH1 was shown to regulate lipid synthesis and the expression of the NF-κB by inhibiting IκB (IκBα) phosphorylation and promoting the nuclear translocation of NF-κB P6553,54, and IDH1 overexpression resulted in increased intracellular α-ketoglutarate levels and downregulation of the PPARγ pathway via the regulation of histone methylation55. These findings suggest a potential crosstalk between IDH1 and the PPARγ pathway. Moreover, we found that the expression of IDH1 was significantly downregulated in pancreatic tissues, pancreatic exosomes, and plasma exosomes of SAP rats after emodin treatment, suggesting that the pancreatic exosomes might regulate the PPARγ/NF-κB pathway of lung tissue via the transfer of a set of proteins (e.g., IDH1) in SAP rats. This is only one of the possible mechanisms by which emodin attenuate SAP-ALI, through the regulation of the secretion of the pancreatic exosomes. Nevertheless, this study provided a direction for future research on the role of exosomes in inflammation-related diseases.
Emodin has been shown to suppress lipopolysaccharide-induced NF-κB activation and inflammation in RAW264.7 mouse macrophages via a PPARγ-dependent mechanism56. Therefore, we further analyzed the changes in the PPARγ/NF-κB signals in alveolar macrophages and lung tissues after the emodin-exos treatment, and we found that emodin-primed exosomes increased PPARγ expression and reduced the levels of p-NF-κB and pro-inflammatory factors in alveolar macrophages and rat lung tissues. These findings were consistent with the previous reports that the PPARγ/NF-κB signaling pathway affects the polarization of macrophages and the secretion of pro-inflammatory factors57, 58, 59. Our results suggest that the PPARγ-NF-κB pathway is an important target of SAP exosomes in SAP-ALI, while this regulation can be suppressed by emodin. Rosiglitazone is a classic PPARγ agonist that inhibits the phosphorylation of PPARγ at the residue S11237,40,41,57. Consistent with previous reports41,60, we found that rosiglitazone inhibited the expression of p-PPARγ in alveolar macrophages and lung tissues at the residue S112, increased the expression of total PPARγ in alveolar macrophages, but had no effect on the expression of total PPARγ in lung tissues in healthy rats. The effects of rosiglitazone on PPARγ expression varied in different cells and/or conditions. For example, rosiglitazone did not affect the expression of total PPARγ in 3T3-L1 adipocytes61, while rosiglitazone increased the expression of total PPARγ in the airway tissue of the asthmatic mouse model39, and rosiglitazone was found to up-regulate the expression of total PPARγ protein in the background of paraquat or LPS treatment38,62. Therefore, the mechanisms by which rosiglitazone upregulated PPARγ expression in alveolar macrophages while not affecting PPARγ expression in the lung tissue need to be explored further. In line with the findings of other studies, that rosiglitazone-induced reduction in inflammation was mediated via the downregulation of the NF-κB pathway63,64. We also found that rosiglitazone-induced reduction in lung inflammation was mediated via the downregulation of p-NF-κB in alveolar macrophages and lung tissues. Altogether, our results suggested that emodin-exos restored PPARγ and inhibited the NF-κB pathway and cytokine secretion in alveolar macrophages and lung tissues of SAP. To further verify that exosomes are involved in the inflammatory regulation of alveolar macrophages and emodin reduces this inflammation by regulating exosomes, GW4869 was used to block the exosomes production in acinar cells. As a result, the anti-inflammatory effect of supernatants from the emodin treatment group was largely reduced by GW4869, suggesting that emodin reduced inflammation of alveolar macrophages partly by regulating the production of exosomes. Furthermore, the plasma exosomes of SAP-ALI rats treated with emodin inhibited the inflammatory NF-κB pathway and reduced the expression of pro-inflammatory factors in the lung tissue of SAP-ALI rats. Taken together, emodin ameliorates SAP-ALI, at least partly by inhibiting the pancreatic exosome-triggered pro-inflammatory signals in alveolar macrophages.
5. Conclusions
We demonstrated that emodin ameliorates SAP-ALI in rats, and that effect is, at least partly dependent on an exosomal mechanism. In brief, emodin reduced harmful pancreatic exosome production and altered the contents of these pathological exosomes under SAP conditions, which consequently suppressed the M1 polarization of the alveolar macrophages and the cytokine release in the lungs, by regulating the PPARγ/NF-κB pathway. In conclusion, this study indicated that blocking pancreatic exosome-mediated alveolar macrophage activation may be a promising strategy to alleviate SAP-ALI, and revealed a vital role of exosomes in participating inflammation associated organ injury.
Acknowledgments
This study was supported by Natural Science Foundation of China (Nos. 81974552, 81774160), Technology Agency of Sichuan Province Foundation Project (No. 2019YFS0389), and Scientific Research Foundation of the Science and Technology Department of Sichuan Province (No. 2022YFS0417, China).
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2021.10.008.
Contributor Information
Wenfu Tang, Email: tangwf@scu.edu.cn.
Jingping Liu, Email: liujingping@scu.edu.cn.
Meihua Wan, Email: wanmh@scu.edu.cn.
Author contributions
Wenfu Tang, Jingping Liu, and Meihua Wan designed that experiment. Qian Hu, Jiaqi Yao and Xiajia Wu conducted the experiments. Qian Hu drafted the main part of the manuscript and analyzed the date. Juan Li and Guixiang Li edited the manuscript. All authors read and approved the final manuscript.
Conflicts of interest
The authors have no conflicts of interest to declare.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Lee P.J., Papachristou G.I. New insights into acute pancreatitis. Nat Rev Gastroenterol Hepatol. 2019;16:479–496. doi: 10.1038/s41575-019-0158-2. [DOI] [PubMed] [Google Scholar]
- 2.Petrov M.S., Yadav D. Global epidemiology and holistic prevention of pancreatitis. Nat Rev Gastroenterol Hepatol. 2019;16:175–184. doi: 10.1038/s41575-018-0087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leppäniemi A., Tolonen M., Tarasconi A., Segovia-Lohse H., Gamberini E., Kirkpatrick A.W., et al. 2019 WSES guidelines for the management of severe acute pancreatitis. World J Emerg Surg. 2019;14:27. doi: 10.1186/s13017-019-0247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wig J.D., Bharathy K.G., Kochhar R., Yadav T.D., Kudari A.K., Doley R.P., et al. Correlates of organ failure in severe acute pancreatitis. Jop. 2009;10:271–275. [PubMed] [Google Scholar]
- 5.Zhu A.J., Shi J.S., Sun X.J. Organ failure associated with severe acute pancreatitis. World J Gastroenterol. 2003;9:2570–2573. doi: 10.3748/wjg.v9.i11.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garg P.K., Singh V.P. Organ failure due to systemic injury in acute pancreatitis. Gastroenterology. 2019;156:2008–2023. doi: 10.1053/j.gastro.2018.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang M., Lei R. Organ dysfunction in the course of severe acute pancreatitis. Pancreas. 2016;45:e5–e7. doi: 10.1097/MPA.0000000000000450. [DOI] [PubMed] [Google Scholar]
- 8.De Campos T., Deree J., Coimbra R. From acute pancreatitis to end-organ injury: mechanisms of acute lung injury. Surg Infect (Larchmt) 2007;8:107–120. doi: 10.1089/sur.2006.011. [DOI] [PubMed] [Google Scholar]
- 9.Manohar M., Verma A.K., Venkateshaiah S.U., Sanders N.L., Mishra A. Chronic pancreatitis associated acute respiratory failure. MOJ Immunol. 2017;5 doi: 10.15406/moji.2017.05.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou M.T., Chen C.S., Chen B.C., Zhang Q.Y., Andersson R. Acute lung injury and ARDS in acute pancreatitis: mechanisms and potential intervention. World J Gastroenterol. 2010;16:2094–2099. doi: 10.3748/wjg.v16.i17.2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akbarshahi H., Rosendahl A.H., Westergren-Thorsson G., Andersson R. Acute lung injury in acute pancreatitis—awaiting the big leap. Respir Med. 2012;106:1199–1210. doi: 10.1016/j.rmed.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 12.Gea-Sorlí S., Guillamat R., Serrano-Mollar A., Closa D. Activation of lung macrophage subpopulations in experimental acute pancreatitis. J Pathol. 2011;223:417–424. doi: 10.1002/path.2814. [DOI] [PubMed] [Google Scholar]
- 13.Shields C.J., Winter D.C., Redmond H.P. Lung injury in acute pancreatitis: mechanisms, prevention, and therapy. Curr Opin Crit Care. 2002;8:158–163. doi: 10.1097/00075198-200204000-00012. [DOI] [PubMed] [Google Scholar]
- 14.Dong X., Fu J., Yin X., Cao S., Li X., Lin L., et al. Emodin: a review of its pharmacology, toxicity and pharmacokinetics. Phytother Res. 2016;30:1207–1218. doi: 10.1002/ptr.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monisha B.A., Kumar N., Tiku A.B. Emodin and its role in chronic diseases. Adv Exp Med Biol. 2016;928:47–73. doi: 10.1007/978-3-319-41334-1_3. [DOI] [PubMed] [Google Scholar]
- 16.Zhu L., Li J.Y., Zhang Y.M., Kang H.X., Chen H., Su H., et al. Pharmacokinetics and pharmacodynamics of shengjiang decoction in rats with acute pancreatitis for protecting against multiple organ injury. World J Gastroenterol. 2017;23:8169–8181. doi: 10.3748/wjg.v23.i46.8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miao Y.F., Kang H.X., Li J., Zhang Y.M., Ren H.Y., Zhu L., et al. Effect of sheng-jiang powder on multiple-organ inflammatory injury in acute pancreatitis in rats fed a high-fat diet. World J Gastroenterol. 2019;25:683–695. doi: 10.3748/wjg.v25.i6.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia S., Ni Y., Zhou Q., Liu H., Xiang H., Sui H., et al. Emodin attenuates severe acute pancreatitis via antioxidant and anti-inflammatory activity. Inflammation. 2019;42:2129–2138. doi: 10.1007/s10753-019-01077-z. [DOI] [PubMed] [Google Scholar]
- 19.Yao W.Y., Zhou Y.F., Qian A.H., Zhang Y.P., Qiao M.M., Zhai Z.K., et al. Emodin has a protective effect in cases of severe acute pancreatitis via inhibition of nuclear factorkappaB activation resulting in antioxidation. Mol Med Rep. 2015;11:1416–1420. doi: 10.3892/mmr.2014.2789. [DOI] [PubMed] [Google Scholar]
- 20.Gao Z., Sui J., Fan R., Qu W., Dong X., Sun D. Emodin protects against acute pancreatitis-associated lung injury by inhibiting NLPR3 inflammasome activation via Nrf2/HO-1 signaling. Drug Des Dev Ther. 2020;14:1971–1982. doi: 10.2147/DDDT.S247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui H., Li S., Xu C., Zhang J., Sun Z., Chen H. Emodin alleviates severe acute pancreatitis-associated acute lung injury by decreasing pre-B-cell colony-enhancing factor expression and promoting polymorphonuclear neutrophil apoptosis. Mol Med Rep. 2017;16:5121–5128. doi: 10.3892/mmr.2017.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu C., Zhang J., Liu J., Li Z., Liu Z., Luo Y., et al. Proteomic analysis reveals the protective effects of emodin on severe acute pancreatitis induced lung injury by inhibiting neutrophil proteases activity. J Proteomics. 2020;220:103760. doi: 10.1016/j.jprot.2020.103760. [DOI] [PubMed] [Google Scholar]
- 23.Dang S.C., Jiang D.L., Chen M., Li D., Zhang J.X. Clodronate-containing liposomes attenuate lung injury in rats with severe acute pancreatitis. J Zhejiang Univ - Sci B. 2010;11:828–835. doi: 10.1631/jzus.B1000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lanyu Z., Feilong H. Emerging role of extracellular vesicles in lung injury and inflammation. Biomed Pharmacother. 2019;113:108748. doi: 10.1016/j.biopha.2019.108748. [DOI] [PubMed] [Google Scholar]
- 25.Lee H., Abston E., Zhang D., Rai A., Jin Y. Extracellular vesicle: an emerging mediator of intercellular crosstalk in lung inflammation and injury. Front Immunol. 2018;9:924. doi: 10.3389/fimmu.2018.00924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun K., He S.B., Qu J.G., Dang S.C., Chen J.X., Gong A.H., et al. IRF5 regulates lung macrophages M2 polarization during severe acute pancreatitis in vitro. World J Gastroenterol. 2016;22:9368–9377. doi: 10.3748/wjg.v22.i42.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu Q., Su H., Li J., Lyon C., Tang W., Wan M., et al. Clinical applications of exosome membrane proteins. Precis Clin Med. 2020;3:54–66. doi: 10.1093/pcmedi/pbaa007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wan M., Ning B., Spiegel S., Lyon C.J., Hu T.Y. Tumor-derived exosomes (TDEs): how to avoid the sting in the tail. Med Res Rev. 2020;40:385–412. doi: 10.1002/med.21623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jimenez-Alesanco A., Marcuello M., Pastor-Jimenez M., Lopez-Puerto L., Bonjoch L., Gironella M., et al. Acute pancreatitis promotes the generation of two different exosome populations. Sci Rep. 2019;9:19887. doi: 10.1038/s41598-019-56220-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonjoch L., Casas V., Carrascal M., Closa D. Involvement of exosomes in lung inflammation associated with experimental acute pancreatitis. J Pathol. 2016;240:235–245. doi: 10.1002/path.4771. [DOI] [PubMed] [Google Scholar]
- 31.Jiang K., Yang J., Guo S., Zhao G., Wu H., Deng G. Peripheral circulating exosome-mediated delivery of miR-155 as a novel mechanism for acute lung inflammation. Mol Ther. 2019;27:1758–1771. doi: 10.1016/j.ymthe.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X.B., Sun H.Y., Luo Z.L., Cheng L., Duan X.M., Ren J.D. Plasma-derived exosomes contribute to pancreatitis-associated lung injury by triggering NLRP3-dependent pyroptosis in alveolar macrophages. Biochim Biophys Acta Mol Basis Dis. 2020;1866:165685. doi: 10.1016/j.bbadis.2020.165685. [DOI] [PubMed] [Google Scholar]
- 33.Hu Q., Lyon C.J., Fletcher J.K., Tang W., Wan M., Hu T.Y. Extracellular vesicle activities regulating macrophage- and tissue-mediated injury and repair responses. Acta Pharm Sin B. 2021;11:1493–1512. doi: 10.1016/j.apsb.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y.M., Ren H.Y., Zhao X.L., Li J., Li J.Y., Wu F.S., et al. Pharmacokinetics and pharmacodynamics of Da-Cheng-Qi decoction in the liver of rats with severe acute pancreatitis. World J Gastroenterol. 2017;23:1367–1374. doi: 10.3748/wjg.v23.i8.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt J., Rattner D.W., Lewandrowski K., Compton C.C., Mandavilli U., Knoefel W.T., et al. A better model of acute pancreatitis for evaluating therapy. Ann Surg. 1992;215:44–56. doi: 10.1097/00000658-199201000-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikawa K., Nishina K., Takao Y., Obara H. ONO-1714, a nitric oxide synthase inhibitor, attenuates endotoxin-induced acute lung injury in rabbits. Anesth Analg. 2003;97:1751–1755. doi: 10.1213/01.ANE.0000086896.90343.13. [DOI] [PubMed] [Google Scholar]
- 37.Ribeiro Filho H.V., Guerra J.V., Cagliari R., Batista F.A.H., Le Maire A., Oliveira P.S.L., et al. Exploring the mechanism of PPARgamma phosphorylation mediated by CDK5. J Struct Biol. 2019;207:317–326. doi: 10.1016/j.jsb.2019.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H., You L., Zhao M. Rosiglitazone attenuates paraquat-induced lung fibrosis in rats in a PPAR gamma-dependent manner. Eur J Pharmacol. 2019;851:133–143. doi: 10.1016/j.ejphar.2019.02.037. [DOI] [PubMed] [Google Scholar]
- 39.Xu J., Zhu Y.T., Wang G.Z., Han D., Wu Y.Y., Zhang D.X., et al. The PPARgamma agonist, rosiglitazone, attenuates airway inflammation and remodeling via heme oxygenase-1 in murine model of asthma. Acta Pharmacol Sin. 2015;36:171–178. doi: 10.1038/aps.2014.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolli V., Stechschulte L.A., Dowling A.R., Rahman S., Czernik P.J., Lecka-Czernik B. Partial agonist, telmisartan, maintains PPARgamma serine 112 phosphorylation, and does not affect osteoblast differentiation and bone mass. PLoS One. 2014;9 doi: 10.1371/journal.pone.0096323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stechschulte L.A., Czernik P.J., Rotter Z.C., Tausif F.N., Corzo C.A., Marciano D.P., et al. PPARG post-translational modifications regulate bone formation and bone resorption. EBioMedicine. 2016;10:174–184. doi: 10.1016/j.ebiom.2016.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beninson L.A., Fleshner M. Exosomes: an emerging factor in stress-induced immunomodulation. Semin Immunol. 2014;26:394–401. doi: 10.1016/j.smim.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Baixauli F., Lopez-Otin C., Mittelbrunn M. Exosomes and autophagy: coordinated mechanisms for the maintenance of cellular fitness. Front Immunol. 2014;5:403. doi: 10.3389/fimmu.2014.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ostrowski M., Carmo N.B., Krumeich S., Fanget I., Raposo G., Savina A., et al. RAB27A and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19–30. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 45.Xu J., Camfield R., Gorski S.M. The interplay between exosomes and autophagy—partners in crime. J Cell Sci. 2018;131:jcs215210. doi: 10.1242/jcs.215210. [DOI] [PubMed] [Google Scholar]
- 46.Closa D., Sabater L., Fernández-Cruz L., Prats N., Gelpí E., Roselló-Catafau J. Activation of alveolar macrophages in lung injury associated with experimental acute pancreatitis is mediated by the liver. Ann Surg. 1999;229:230–236. doi: 10.1097/00000658-199902000-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neri T., Armani C., Pegoli A., Cordazzo C., Carmazzi Y., Brunelleschi S., et al. Role of NF-kappaB and PPAR-gamma in lung inflammation induced by monocyte-derived microparticles. Eur Respir J. 2011;37:1494–1502. doi: 10.1183/09031936.00023310. [DOI] [PubMed] [Google Scholar]
- 48.Velickovic K., Lugo Leija H.A., Surrati A., Kim D.H., Sacks H., Symonds M.E., et al. Targeting glutamine synthesis inhibits stem cell adipogenesis in vitro. Cell Physiol Biochem. 2020;54:917–927. doi: 10.33594/000000278. [DOI] [PubMed] [Google Scholar]
- 49.Yang R.X., Pan Q., Liu X.L., Zhou D., Xin F.Z., Zhao Z.H., et al. Therapeutic effect and autophagy regulation of myriocin in nonalcoholic steatohepatitis. Lipids Health Dis. 2019;18:179. doi: 10.1186/s12944-019-1118-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Z., Zhang H.Y., Zhang Y., Li H. Inactivation of the Ras/MAPK/PPARγ signaling axis alleviates diabetic mellitus-induced erectile dysfunction through suppression of corpus cavernosal endothelial cell apoptosis by inhibiting HMGCS2 expression. Endocrine. 2019;63:615–631. doi: 10.1007/s12020-018-1810-2. [DOI] [PubMed] [Google Scholar]
- 51.Simula M.P., Cannizzaro R., Canzonieri V., Pavan A., Maiero S., Toffoli G., et al. PPAR signaling pathway and cancer-related proteins are involved in celiac disease-associated tissue damage. Mol Med. 2010;16:199–209. doi: 10.2119/molmed.2009.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han S., Liu Y., Cai S.J., Qian M., Ding J., Larion M., et al. IDH mutation in glioma: molecular mechanisms and potential therapeutic targets. Br J Cancer. 2020;122:1580–1589. doi: 10.1038/s41416-020-0814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang G., Sai K., Gong F., Yang Q., Chen F., Lin J. Mutation of isocitrate dehydrogenase 1 induces glioma cell proliferation via nuclear factor-kappaB activation in a hypoxia-inducible factor 1-alpha dependent manner. Mol Med Rep. 2014;9:1799–1805. doi: 10.3892/mmr.2014.2052. [DOI] [PubMed] [Google Scholar]
- 54.Liu X., Sun H., Yu M., Liu J., Yang B., Wu Y., et al. DDIT3 regulates cementoblast mineralization by isocitrate dehydrogenase 1 through nuclear factor-kappaB pathway. J Cell Physiol. 2019;234:11602–11609. doi: 10.1002/jcp.27811. [DOI] [PubMed] [Google Scholar]
- 55.Kang H.S., Lee J.H., Oh K.J., Lee E.W., Han B.S., Park K.Y., et al. IDH1-dependent alpha-KG regulates brown fat differentiation and function by modulating histone methylation. Metabolism. 2020;105:154173. doi: 10.1016/j.metabol.2020.154173. [DOI] [PubMed] [Google Scholar]
- 56.Zhu T., Zhang W., Feng S.J., Yu H.P. Emodin suppresses LPS-induced inflammation in RAW264.7 cells through a PPARγ-dependent pathway. Int Immunopharmacol. 2016;34:16–24. doi: 10.1016/j.intimp.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 57.Burns K.A., Vanden Heuvel J.P. Modulation of PPAR activity via phosphorylation. Biochim Biophys Acta. 2007;1771:952–960. doi: 10.1016/j.bbalip.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo W., Xu Q., Wang Q., Wu H., Hua J. Effect of modulation of PPAR-gamma activity on Kupffer cells M1/M2 polarization in the development of non-alcoholic fatty liver disease. Sci Rep. 2017;7:44612. doi: 10.1038/srep44612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nelson V.L., Nguyen H.C.B., Garcìa-Cañaveras J.C., Briggs E.R., Ho W.Y., DiSpirito J.R., et al. PPARγ is a nexus controlling alternative activation of macrophages via glutamine metabolism. Gene Dev. 2018;32:1035–1044. doi: 10.1101/gad.312355.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu Y.D., Yu S.L., Wang R., Liu J.N., Jin Y.S., Li Y.F., et al. Rosiglitazone suppresses calcium oxalate crystal binding and oxalate-induced oxidative stress in renal epithelial cells by promoting PPAR-γ activation and subsequent regulation of TGF-β1 and HGF expression. Oxid Med Cell Longev. 2019;2019:4826525. doi: 10.1155/2019/4826525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choi J.H., Banks A.S., Estall J.L., Kajimura S., Bostrom P., Laznik D., et al. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARgamma by Cdk5. Nature. 2010;466:451–456. doi: 10.1038/nature09291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ji X.X., Ji X.J., Li Q.Q., Lu X.X., Luo L. Rosiglitazone reduces apoptosis and inflammation in lipopolysaccharide-induced human umbilical vein endothelial cells. Med Sci Monit. 2018;24:6200–6207. doi: 10.12659/MSM.910036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kadam L., Kilburn B., Baczyk D., Kohan-Ghadr H.R., Kingdom J., Drewlo S. Rosiglitazone blocks first trimester in-vitro placental injury caused by NF-kappaB-mediated inflammation. Sci Rep. 2019;9:2018. doi: 10.1038/s41598-018-38336-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y.F., Zou X.L., Wu J., Yu X.Q., Yang X. Rosiglitazone, a peroxisome proliferator-activated receptor (PPAR)-gamma agonist, attenuates inflammation via NF-kappaB inhibition in lipopolysaccharide-induced peritonitis. Inflammation. 2015;38:2105–2115. doi: 10.1007/s10753-015-0193-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.