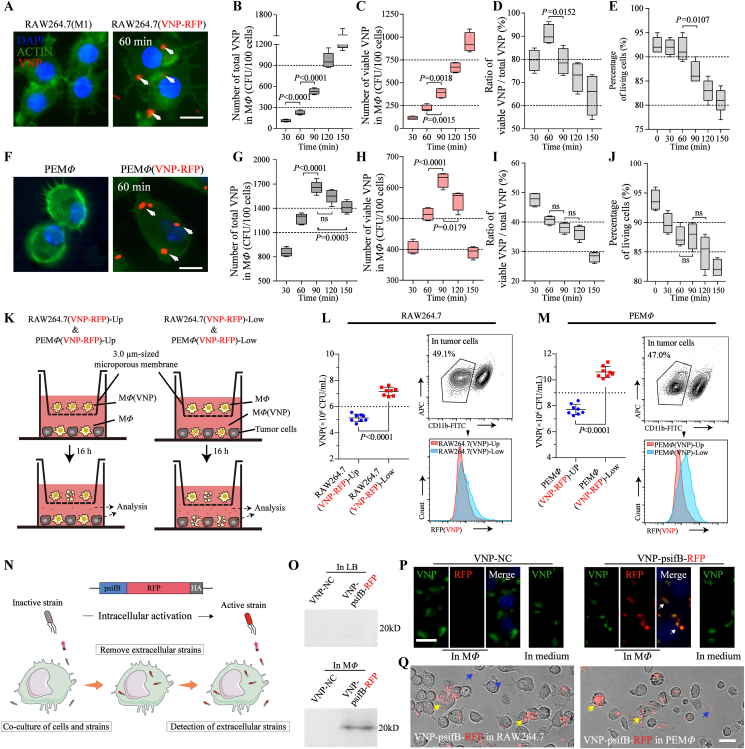
Figure 1.
Preparation of MΦ(VNP) cells and the release of VNP from MΦ(VNP) cells. (A–E) RAW264.7 cells were induced with LPS (100 ng/mL, 12 h) and cocultured with VNP-RFP (1:10) for different times (30–150 min). Gentamicin (50 μg/mL) was added to kill extracellular VNP-RFP, and the quantity of the loaded bacteria in cells and cell viability were examined (n = 5). (A) Fluorescent images of the loaded cells. RAW264.7 cells were cocultured with VNP-RFP for 60 min and stained with DAPI and actin–FITC (Scale bar = 10 μm). (B) Changes in the total number of VNP-RFP strains phagocytosed by RAW264.7 cells. (C) Changes in the number of surviving VNP-RFP strains phagocytosed by RAW264.7 cells after different coculture times. (D) Changes in the loading efficiency (number of live bacteria/number of total bacteria) in macrophages. (E) The percentage of living RAW264.7 cells after different coculture times. (F–J) The results of repeating the experiments in (A–E) by replacing RAW264.7 cells with peritoneal macrophages (PEMΦ) (n = 5). (K–M) Detection of VNP-RFP released from MΦ(VNP-RFP) cells. (K) RAW264.7(VNP-RFP) or PEMΦ(VNP-RFP) (40,000 cells) cells were cultured in medium supplemented with 50 μg/mL gentamicin for 12 h, and then gentamicin was removed and cultured with B16F10 cells indirectly (RAW264.7(VNP-RFP)-Up & PEMΦ(VNP-RFP)-Up) or directly (RAW264.7(VNP-RFP)-Low & PEMΦ(VNP-RFP)-Low) by using a 3.0-μm Transwell chamber. The quantity of VNP in the supernatant or inside tumor cells was determined after 16 h. (L, M) The VNP titer in the supernatant (left) and comparison of the population of VNP inside tumor cells (right). (N) Schematic diagram of the use of VNP-psifB-RFP to examine bacterial release by macrophages. Due to the psifB promoter, VNP-sifB-RFP can only be activated and express RFP intracellularly, so extracellular red bacteria must have escaped from macrophages. (O) Western blot showing RFP expression of VNP-psifB-RFP in LB with OD600 = 0.6–0.8 or inside macrophages for 60 min. A strain carrying empty plasmid (VNP-NC) was used as a control. (P) Fluorescent images showing that VNP-psifB-RFP specifically expressed RFP within 60 min after entering into macrophages, while the strain cultured in DMEM did not express RFP. The cells were stained with Salmonella-specific antibodies (green) and DAPI (blue). (Scale bar = 10 μm). (Q) Brightfield images showing activated VNP-psifB-RFP inside RAW264.7 and PEMΦ (yellow arrows) cells and the VNP released from macrophages (blue arrows) after 3 h of culture in medium containing 50 μg/mL gentamicin (Scale bar = 20 μm). The data in (L) and (M) are reported as the mean ± SEM. Boxplot representations of the spot counts. The median, interquartile range, and minimum and maximum identifiers are shown. All data are representative of three independent experiments. Statistics were calculated using the two-tailed, unpaired Student's t test with Welch's correction.