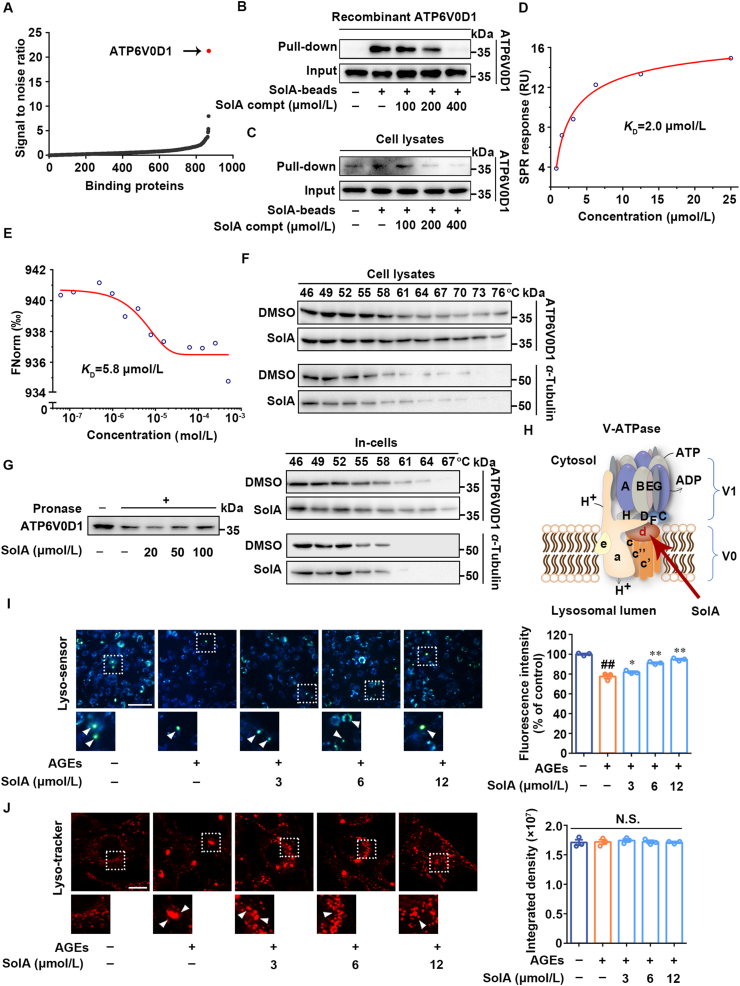
Figure 3.
Identification of V-ATPase subunit ATP6V0D1 as a cellular target of SolA. (A) Identification of cellular target of SolA in PC12 cells proteome. ATP6V0D1 showed the highest signal to noise ratio. (B) SolA selectively bound to recombinant ATP6V0D1 by pull-down analysis. (C) SolA selectively bound to endogenous ATP6V0D1 by pull-down analysis. (D) Binding affinity analysis of SolA with ATP6V0D1 determined by surface plasmon resonance (SPR). (E) Binding affinity analysis of SolA with ATP6V0D1 determined by microscale thermophoresis (MST). (F) SolA promoted ATP6V0D1 resistant to different temperature gradients by cellular thermal shift assays (CETSA) analysis. (G) SolA promoted ATP6V0D1 resistant to proteases by drug affinity responsive target stability (DARTS) analysis. (H) Structure of lysosomal proton pump V-ATPase. (I) SolA promoted lysosomal pH decrease in AGEs-induced PC12 cells (scale bar: 100 μm, n = 3). Arrows indicate highly acidic lysosomes. (J) SolA reversed aberrant lysosomal aggregation in AGEs-induced PC12 cells (scale bar: 10 μm, n = 3). Arrows indicate significantly aggregated lysosomes. Data are expressed as the mean ± SEM. ##P < 0.01 vs. control group, ∗P < 0.05, ∗∗P < 0.01 vs. AGEs group. N.S. not significant.