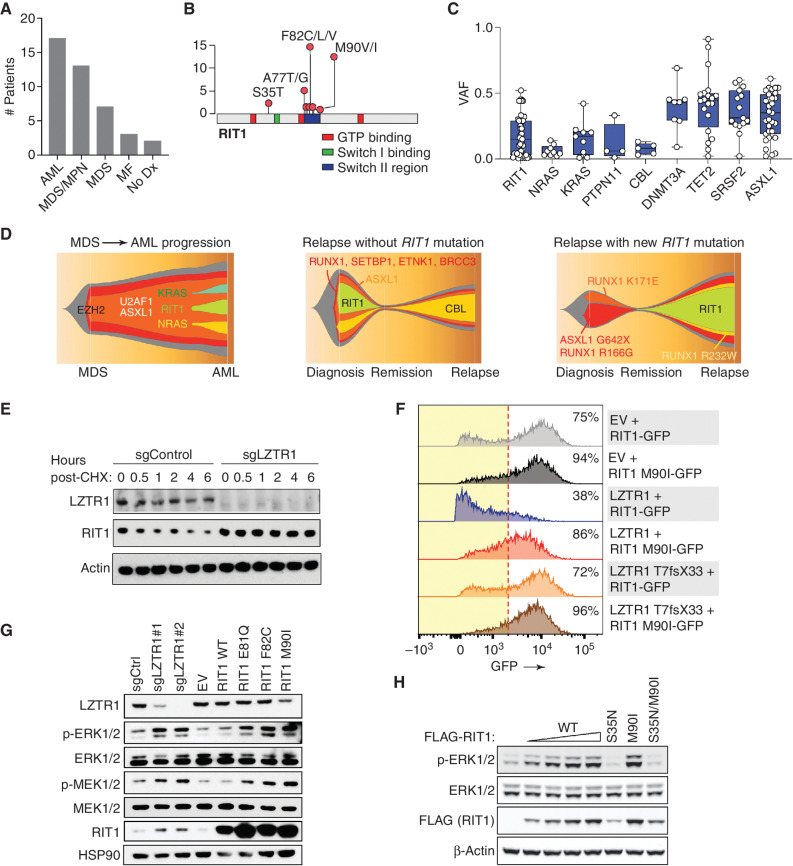
Figure 2.
Characterization of RIT1 mutations in patients with hematologic malignancies. A, Histogram of the number of patients with RIT1 mutations based on myeloid malignancy diagnosis. MF, myelofibrosis; No Dx, no diagnosis. B, Diagram of the location of RIT1 mutations identified. C, Variant allele frequency (VAF) of mutations in RAS GTPases or regulators of RAS-GTP abundance relative to mutations in transcriptional modifiers in patients with myeloid leukemia. D, Fishtail representation plots of VAFs of mutations across a serial genomic analysis of three patients with RIT1 mutations. E, Western blot of LZTR1 and RIT1 following cycloheximide (CHX) treatment of TF-1 cells with or without LZTR1 deletion. sg, single guide. F, Representative histograms of GFP in cells encoding WT or mutant RIT1 fused to eGFP along with empty vector (EV), WT LZTR1, or mutant LZTR1. The percentage of eGFP+ cells is indicated. The red dotted line indicates the cutoff for GFP+. G, Levels of phosphorylated and total MEK1/2 and ERK1/2 as well as RIT1 in TF-1 cells with LZTR1 deletion or expression of EV RIT1 WT or mutant cDNAs. H, Western blot of p-ERK and total ERK levels in 293T cells transfected with increasing amounts of FLAG-RIT1 WT and mutant cDNAs.