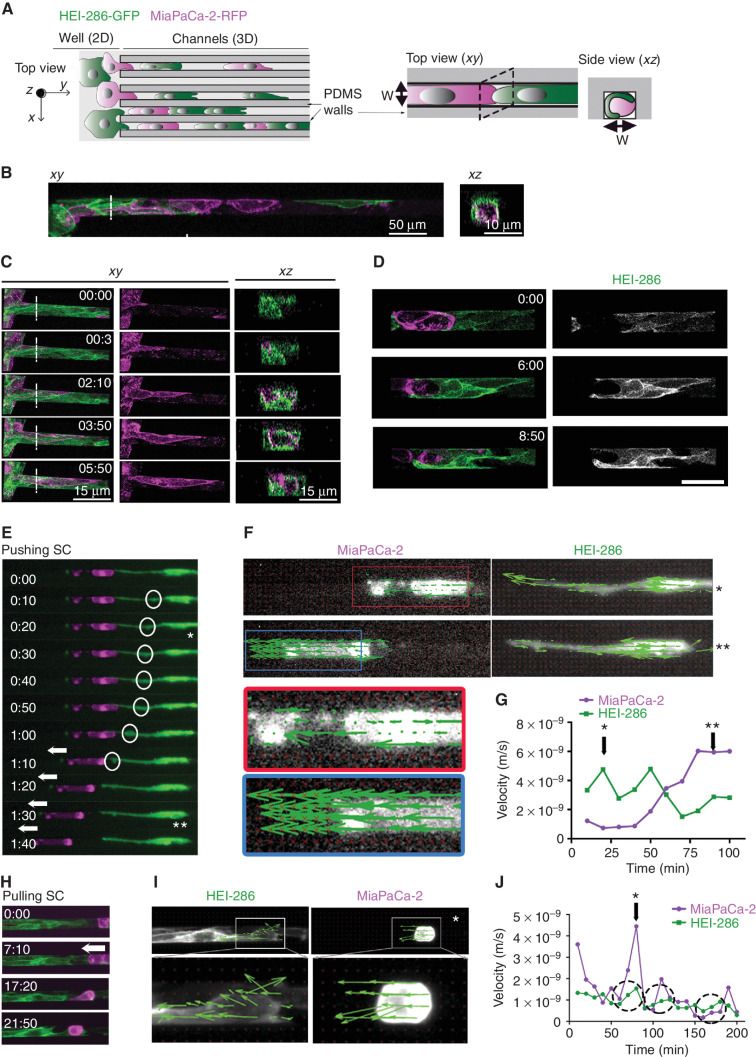
Figure 3.
SCs form dynamic tracks for cancer cells. A, Schematic of microchannels with HEI-286 SCs (green) and MiaPaCa-2 cells (magenta). Both cell types are seeded in the adjacent well (left, 2D) and enter microchannels (3D) where they make contact with each other. PDMS, polydimethylsiloxane. B, Confocal images of HEI-286 SCs and MiaPaCa-2 within microchannels in longitudinal (xy) and transverse (xz) sections. Scale bars, 50 μm. C, Confocal images of time-lapse movies showing a cancer cell moving in a microchannel lined by HEI-286 SCs. Time is h:min. Scale bars, 15 μm. D, Confocal images of time-lapse movies showing an HEI-286 SC wrapping around a cancer cell. Scale bar, 50 μm. E, Fluorescent images of time-lapse movie showing an HEI-286 SC pushing a cancer cell. Arrows indicate cancer cell displacement. Circles indicate intracellular movement within the SC (* and ** are time points shown in E). F, Fluorescent images of an HEI-286 SC and cancer cell from E at two time points (* and **) overlaid with vectors obtained by PIV analysis indicating vector direction. G, Quantification of the mean instantaneous velocity of the HEI-286 SC and cancer cell in E, F, and G. Corresponding time points are indicated by * and **. H, Fluorescent images of time-lapse movie showing HEI-286 SCs pulling a cancer cell. Arrows indicate cancer cell displacement. I, Fluorescent images of the HEI-286 SCs and cancer cells from H at one time point (*) overlaid with vectors obtained by PIV analysis. J, Quantification of the mean instantaneous velocity. *, Corresponding time point in I. Dotted circles indicate periods with synchronized increases in velocity for both cancer cells and SCs.